
THE CRYSTAL STRUCTURE OF GSXYN52B2 SUGGESTS A STRUCTURAL BASIS FOR ITS UNIQUE GLYCOSYNTHASE ACTIVITY
2Department of Biotechnology and Food Engineering, Technion-Israel Institute of Technology, Haifa
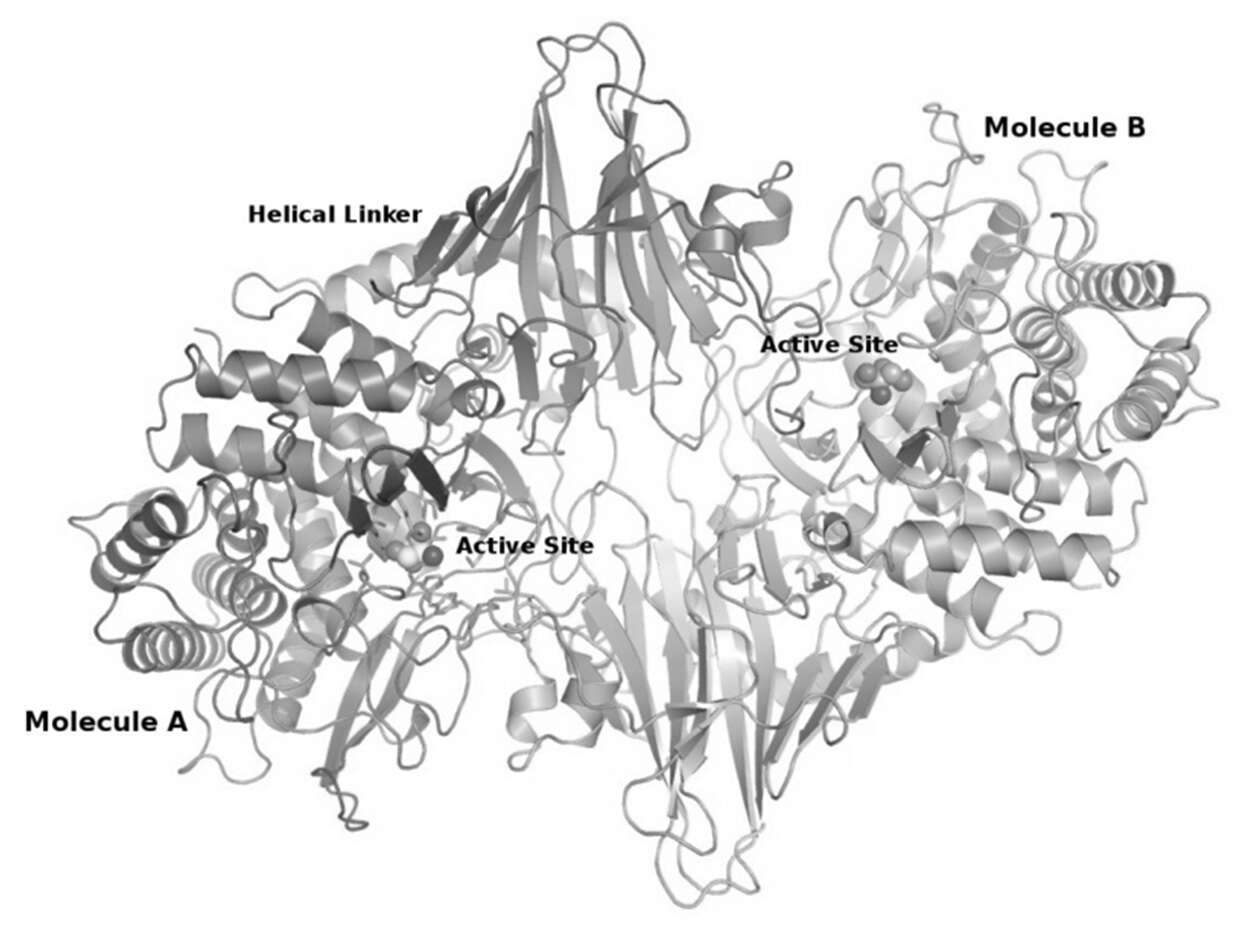
Conversion of glycosyl hydrolases (GHs) into glycosynthases provides an attractive approach for synthesizing carbohydrates. GsXyn52B2, a retaining GH52 b-xylosidase from Geobacillus stearothermophilus, displays significant glycosynthase ability upon mutation of the nucleophile E335 into G335[1]. This activity improves by about 35-fold upon introduction of 10 mutations, obtained through two cycles of directed-evolution experiments[2]. As a first step towards understanding the effect of these mutations, we report here the X-ray crystal structure of the GsXyn52B2-E335G mutant[3], the second 3D protein structure to be solved from the GH52 family. The GsXyn52B2 monomer consists of two domains, a catalytic (α/α)6 barrel domain and a β sandwich domain, connected by a helical linker. Two such GsXyn52B2 monomers interact in a "head-to-tail" fashion to form a biologically functional dimer, as confirmed by gel-filtration, SAXS and theoretical calculations. Tris molecules, resulting from the crystallization buffer, were found trapped in the active sites, and allow identification of the glycon and aglycon binding sites of this enzyme. The current GsXyn52B2-E335G structure enables the structural mapping of the directed-evolution mutations, providing first insights into the structural basis for the enhanced glycosynthase activity of the enzyme mutant and suggesting the key relevant mutations. This information can potentially be used as a basis for the rational mutagenesis of this and related GHs into efficient glycosynthases, and is thus of significant biotechnological importance.
[1] A. Ben-David, T. Bravman, Y.S. Balazs, M. Czjzek, D. Schomburg, G. Shoham, Y. Shoham, ChemBioChem 2007, 8, 2145–2151.
[2] A. Ben-David, G. Shoham, Y. Shoham, Chem. Biol. 2008, 15, 546–551.
[3] R. Dann, S. Lansky, N. Lavid, A. Zehavi, V. Belakhov, T. Baasov, H. Dvir, B. Manjasetty, H. Belrhali, Y. Shoham, G. Shoham, Acta Crystallogr. F, 2014, 70, 1675–82.
Powered by Eventact EMS