
TOWARDS UNDERSTANDING SUMOYLATION: TOTAL CHEMICAL SYNTHESIS OF THE SUMO2 PROTEIN
Although post-translational modifications of proteins expressed in the cell is a widely-studied field, very little is known about the small ubiquitin-like modifier, SUMO. Misregualted SUMO production has been shown to have numerous negative effects, including neurodegenerative disease, diabetes, and tumorigenesis, to name a few.
Nonetheless, very little is understood about the role that SUMO plays in healthy cell function. The reason for this is twofold. Firstly, unlike its counterpart ubiquitin, SUMO is degraded rapidly and is difficult to study under typical in vivo conditions. Secondly, unlike ubiquitin, SUMO is capable of forming longer and heterogeneous chains in the cell. There is no known method for controlling the length of SUMO-chain and so expression of homogeneous SUMO multimers is virtually impossible. For these two reasons, we believe it is essential to develop a robust method for SUMOylation and di-SUMOylation using chemical protein synthesis.
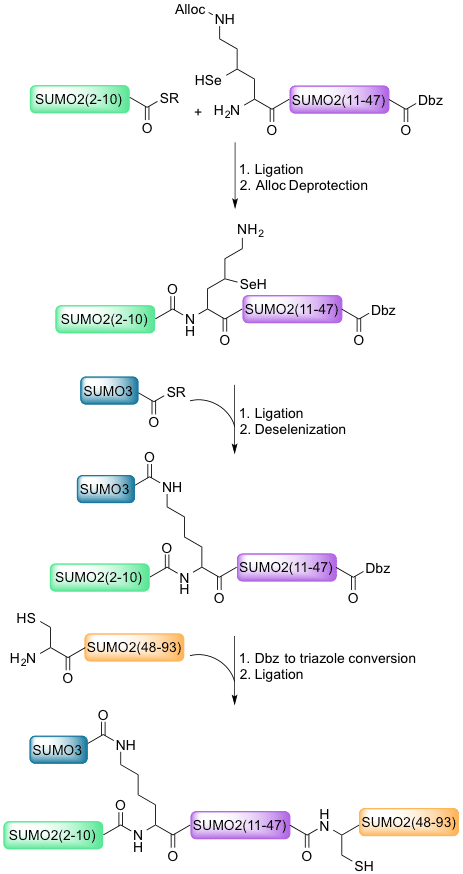
In this project, we are working towards synthesis of di-SUMOylated proteins using common methods in chemical protein synthesis: solid-phase peptide synthesis (SPPS) and native chemical ligation (NCL). Use of gamma-selenolysine will allow for dual chemical ligation at Lys11, the site of chain-forming in SUMOylation. Subsequent selective deselenization using TCEP will leave the first chemically synthesized diSUMO containing only native bonds.
Here we present the first step in our synthesis: the total synthesis of SUMO2. This 93-amino acid chain synthesis serves as a proof-of-concept that gamma-selenolysine is a powerful new amino acid in the NCL toolbox. With this method, we hope to enable the study of SUMO and SUMO multimers’ post-translational effects.
Powered by Eventact EMS