
COVALENT DOCKING OF LARGE VIRTUAL ELECTROPHILE LIBRARIES DISCOVERS NEW POTENT AND SELECTIVE INHIBITORS
2Cellular and Molecular Pharmacology, University of California San Francisco
3Pharmaceutical Chemistry, University of California San Francisco
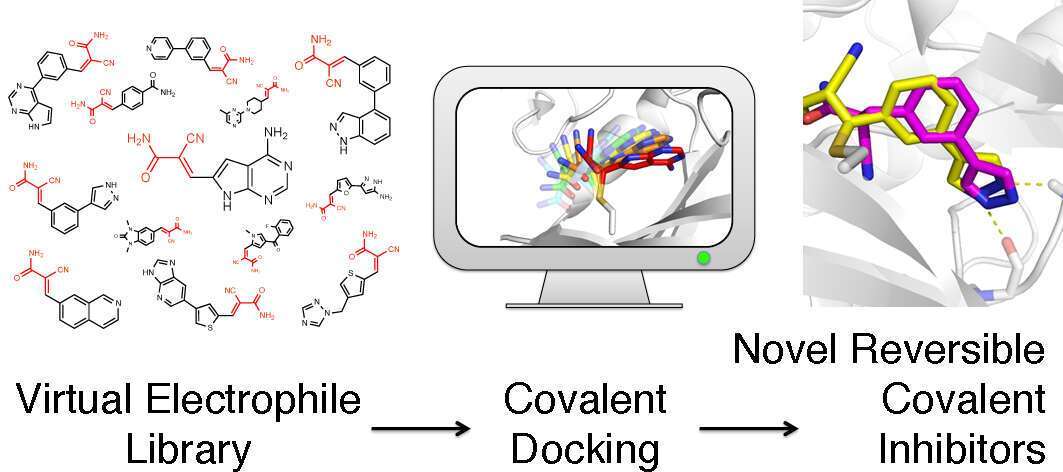
Small molecules are invaluable tools for the investigation of biology. However, discovering new molecules to specifically modulate a target protein is still one of the biggest challenges of chemical biology. Molecules that are able to form a covalent bond with their target often show enhanced selectivity, potency and utility for biological studies, but are yet harder to discover, as they are typically expunged from high throughput screening libraries. Computational methods can help bridge this gap. We developed a covalent docking method for the discovery of covalent probes. By constructing large virtual libraries of synthetically accessible electrophiles and applying this method prospectively to several protein targets we were able to discover potent covalent inhibitors (typically with <50nM IC50), with chemotypes not previously explored. The docking predictions were confirmed by crystallography, the inhibitors displayed marked selectivity and were active in cellular assays. This approach should be applicable for a broad range of protein targets, and many different electrophile chemistries.
Powered by Eventact EMS