
USING CHEMICAL PROTEOMICS TO UNRAVEL THE P. AERUGINOSA QUORUM SENSING REGULON
In recent years the world has seen a troubling increase in antibiotic resistance among many kinds of bacteria, especially in hospitals. Certain pathogen bacteria have gained resistance against most known antibiotics. An alternative approach to treat and / or prevent bacterial infections is to target the bacterial communication mechanisms that enable bacteria to collaborate with one another.
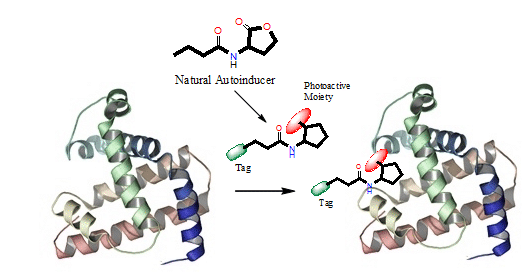
Many bacterial species use a form of chemical communication in order to coordinate gene expression and this system is termed `Quorum Sensing` (QS). This type of cell to cell communication helps the bacteria to sense the population density and express genes in a collective manner. In order to sense their `quorum`, the bacteria use small molecules called autoinducers. These molecules are synthesized by the bacteria and secreted into their surroundings, followed by reception and translation by other bacteria, and thus function as a bacterial "language". This enables the bacteria to assess their population density and regulate virulence accordingly. Our aim is to attain a deep molecular understanding of these processes through synthesis of modified autoinducers. By using such probes we aim to target specific receptors and also discover unknown proteins that are part of the QS regulon.
In Pseudomonas aeruginosa one of the native autoinducers is N-butyryl-L-homoserine lactone (C4-HSL). We designed and modified the basic structure of this native autoinducer and introduced a covalently binding photoactive group and a ‘click’ tag as an analytical handle. Using an activity based protein profiling (ABPP) platform we will be able to identify unknown C4-HSL binding proteins and further unravel the full P. aeruginosa QS regulon.
Powered by Eventact EMS