Paramecium Bursaria Chlorella Virus-1 (PBCV-1) is the prototype member of Phycodnaviridae family and shares common features with other group members of the Nucleo Cytoplasmic Large DNA Viruses (NCLDV) clade. The virus is an icosahedral shaped and it harbors ~331 Kbp of dsDNA genome enclosed by one internal membrane. Except for Mimivirus and Vaccinia Virus all DNA viruses replicate their genomes in the nucleus of their hosts. The rationale is that the nucleus is a suitable platform to carryout viral transactions such as DNA replication and transcription of viral genes. In the case of PBCV-1 it was postulated that viral genomes have to be transported to the nucleus of the Chlorella cell in order to enable viral DNA replication and this is due to the facts that no recognizable RNA polymerase gene was identified in the virus genome and that several viral genes possess introns that have to be cleaved by the spliceosome machinery. Trafficking of large genomes in the highly crowded cytoplasmic milieu is associated with substantial barriers including intracellular membranous compartments and cytoplasmic nucleases. In addition, the large chloroplast of the Chlorella cells act as a substantial barrier for the movement of large molecular assemblies. In our studies we investigated the infection cycle of the large dsDNA virus PBCV-1 in details. By combining advanced high-resolution fluorescence and electron microscopy techniques we have shown that in contrast to other members of the NCLDV, PBCV-1 infection is initiated with a unique mechanism reminiscent that of bacteriophages. Using Scanning Transmission Electron Microscopy (STEM) tomography we showed that soon after adsorption to the cell wall, PBCV-1 generates a hole in the cell walls through which a narrow membranous tunnel is formed by the fusion of the plasma and viral membranes, and is used for viral genome delivery into the cytoplasm. We carried out high resolution florescence imaging studies using Stochastic Optical Reconstruction Microscopy (STORM) in order to track viral genomes. These studies revealed that soon after being ejected into the cytoplasm viral DNA is transported to the nucleus. The STORM studies were further supported by immuno-labeling experiments of high pressure frozen and freeze substituted (HPF-FS) Chlorella infected cells and Electron Microscopy In Situ Hybridization (EMISH) assays that provides specific detection and localization of viral DNA. Viral DNA replication most likely occurs in the nucleus of the cells. Our results strongly support this notion as we were able to detect viral DNA egress from nuclei of infected cells in thin TEM sections of HPF-FS Chlorella infected cells that were labeld with DNA antibodies. The infection proceeds with the generation of elaborated "viral organelles", termed, viral factories that are generated in the cytoplasm of the host. These organelles are the sites where viral transactions such as DNA replication, transcription of viral genes, membrane biogenesis, packaging of viral DNA and assembly of new progeny, take place. The viral factories generated by PBCV-1 are profoundly different from those produced by other group members of the NCLDVs, Vaccinia virus, African Swine Fever Virus (ASFV) and Mimivirus. We used several techniques that provide 3D information with nanometer resolution, specifically, STEM tomography and Focused Ion Beam Scanning Electron Microscopy (FIB-SEM), in order to investigate the 3D organization and function of the factories generated by PBCV-1. These experiments were corroborated by florescence microscopy studies. Our studies revealed the distinct rosette-like architecture of the factories, with viral genomes at the outer periphery, host cisternae at the inner periphery, and single bilayer membrane sheets used as capsid templates in the central region. The cisternae bud out from outer nuclear membranes and translocate to the periphery of the factories and even penetrate deep in to the core of the factory were they are rendered into single open membrane sheets. Virus assembly takes place at the periphery of the factory, and at the end of the process, viral DNA is packed through a narrow aperture in a well-defined singular vertex. This mechanism of membrane biogenesis demonstrated for PBCV-1 is similar to other group members of the NCLDVs such as Mimivirus, ASFV and Vaccinia virus. Overall, our results point towards the notion that the eukaryote-infecting PBCV-1 infection initiates and proceeds through a bacteriophage-like infection.
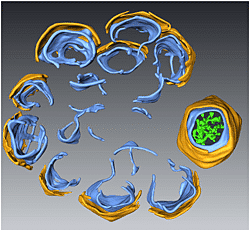
A 3D surface representation of a STEM tomogram showing the rosette-like crescent structure of the viral factories generated by PBCV-1. The crescents are composed of two distinct layers, an external angular capsid (yellow) and an underlying internal membrane (light blue). The core of the factory consists of open membrane sheets that act as templates for the assembly of the capsids.

