INTRODUCTION
Intravital microscopy is a powerful tool, providing the ability to characterize live organism physiology in the natural context of cellular and sub cellular resolution. However the necessity to perform studies on a live organism might associate with severe motion artifacts that should be corrected. Most of the software solutions are dedicated to solve this problem can do it with different levels of success, but they fault in the case when motion artifacts accompanied with a strong structural changes inside the object occur during image acquisition. Here we demonstrate the use of global Kappa-Omega approach to stabilize image without supervision and assignment of a reference image.
METHOD
As an experimental intravital setup we used previously developed Transcranial Optical Vascular Imaging (TOVI) approach [1], where we use a dynamic fluorescence contrast enhanced microscopic images of cerebral blood vessels accompanied with moderate jerks during image acquisition as well as randomly applied larger jerks after post acquisition fig 1.a. Adaptive approach consists in applying the accurate cone shape Kappa-Omega filter on a 3D Fourier transform matrix [2] (that contains mix of all frequencies including spatial and temporal) fig.1b.
RESULTS
Results are presented in a fig. 1c,d in a temporal color-coded manner that clearly demonstrates the efficiency of the adaptive approach by suppressing motion artifacts while saving structural changes during acquisition 1d.
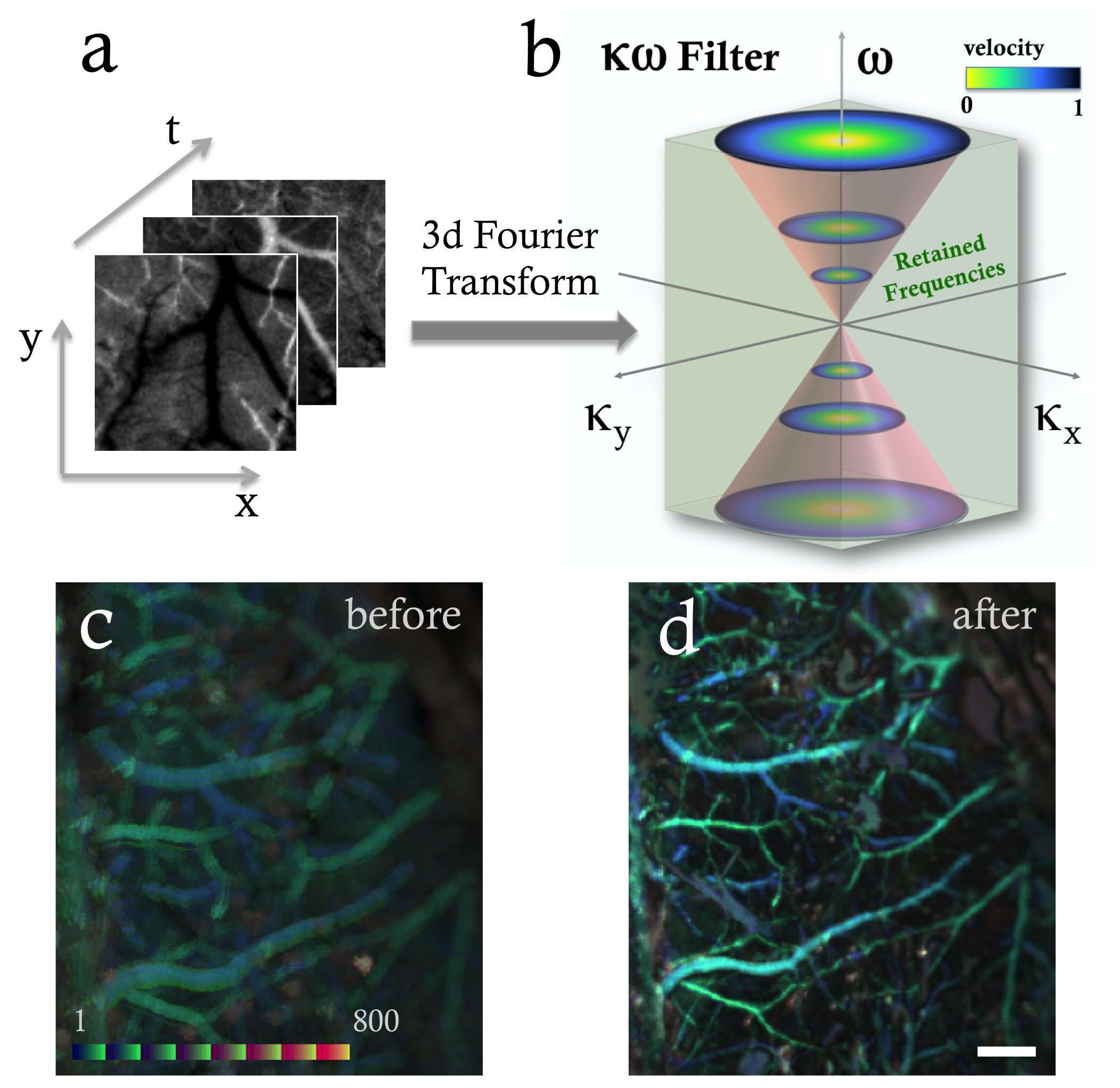
Figure 1.
- a) Represented an image stack x,y of TOVI images over time t.
- b) Time-space Fourier filter represented in the k omega space taking into account the symmetric properties of the Fourier transform.
- c) A temporal color-coded image of cerebral blood vessels through the intact cranium before applying an adaptive filter. Color coded bar is a result of temporal color coding (FIJI / ImageJ) which is applied along the time sequence to distinguish the time evolution structures from the image motions when stacking the 800 images.
- d) A temporal color-coded image after applying an adaptive K-Omega filter (same region of interest). White scale bar is 500um.
CONCLUSIONS
Developed adaptive approach might be useful when alternative techniques fault to be successful to correct moderate motion artifacts associated with structural changes in the image sequence when no reference image can be assigned. Our proposed adaptive approach would be also a useful tool to combine with standard registration method such as correlation methods based on Fourier transforms.
Acknowledgements
Author would like to thank for the partial support of The Henry Chanoch Krenter Institute for Biomedical imaging and Genomics, Staff Scientist program.
References
- Kalchenko, D. Israeli, Y. Kuznetsov, A. Harmelin, “ Transcranial optical vascular imaging (TOVI) of cortical hemodynamics in mouse brain”, Sci Rep. 2014 Jul 25; 4:5839.
- Molodij, E.N. Ribak, M. Glanc, G. Chenegros, “ Enhancing retinal images by nonlinear registration”, Opt. Comm. 342, 157-166 (2015).

