Studying membrane proteins at the nanoscale in intact eukaryotic cells in their native liquid environment has become possible recently using a new approach involving scanning transmission electron microscopy (STEM), termed Liquid STEM [1, 2]. Cells in liquid are placed in a microfluidic chamber enclosing the sample in the vacuum of the electron microscope, and are then imaged with STEM. It is not always necessary to enclose the cells in the microfluidic chamber. For many studies, it is sufficient to obtain information from the thin outer regions of the cells, and those can be imaged with high resolution using environmental scanning electron microscopy (ESEM) with STEM detector [3]. Liquid STEM was used to explore the formation of the epidermal growth factor HER2 at the single-molecule level in intact SKBR3 breast cancer cells in liquid state [4]. HER2 is a membrane protein and plays an important role in breast cancer aggressiveness and progression. Data analysis based on calculating the pair correlation function from individual HER2 positions revealed remarkable differences in functionality between different cellular regions, and between cells with possible relevance for studying cancer metastasis and drug response.
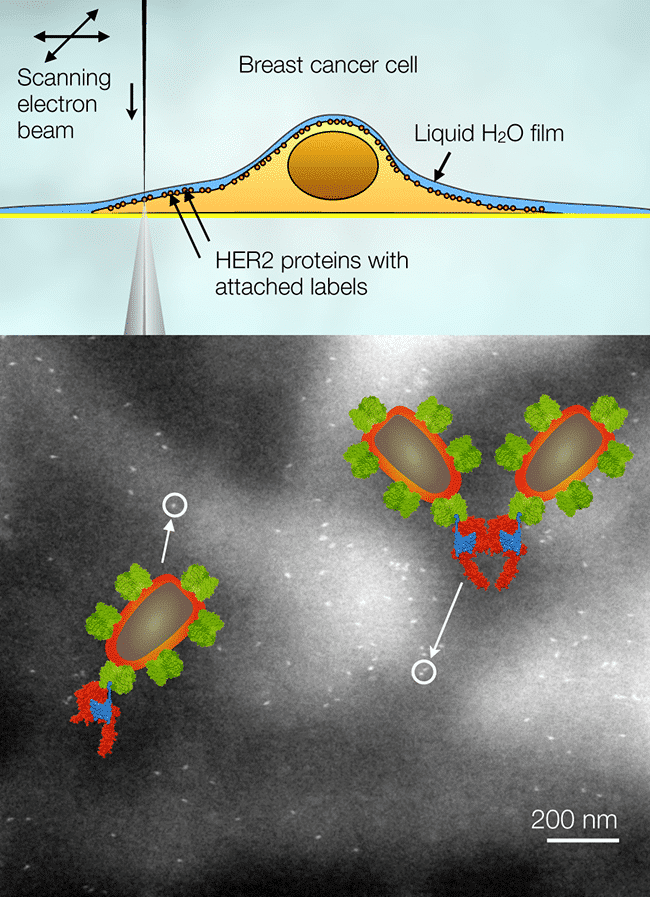
Fig. 1. Electron microscopy of HER2 in whole cancer cells in liquid. (Top) HER2 proteins labeled with nanoparticles within the plasma membrane of a cell are imaged with a scanning electron beam in transmission mode. The cells remain under a thin layer of water. (Bottom) The nanoparticles are visible as bright spots in the images. Individual labels, pairs, and larger groups are found. Two examples are highlighted, a nanoparticle label attached to a HER2 monomer, and two labels attached to a dimer. Colored molecular models are shown as well.
References
[1] de Jonge, N., Peckys, D.B., Kremers, G.J., Piston, D.W., Electron microscopy of whole cells in liquid with nanometer resolution, Proc. Natl. Acad. Sci. 106, 2159-2164, 2009.
[2] de Jonge, N., Ross, F.M., Electron microscopy of specimens in liquid, Nat. Nanotechnol. 6, 695-704, 2011.
[3] Peckys, D.B., Baudoin, J.P., Eder, M., Werner, U., de Jonge, N., Epidermal growth factor receptor subunit locations determined in hydrated cells with environmental scanning electron microscopy, Sci. Rep. 3, 2626: 1-6, 2013.
[4] Peckys, D.B., Korf, U., de Jonge, N., Local variations of HER2 dimerization in breast cancer cells discovered by correlative fluorescence and liquid electron microscopy, Sci. Adv. 1, e1500165, 2015.

