A major challenge for standard electron microscopy has been the identification of rare cellular events or specific cells of interest in a complex tissue. Immuno-EM offers one solution but technical challenges and limitations frequently render this approach insufficient. Correlative Light and Electron Microscopy (CLEM) procedures have been developed to unite optical methods that provide an overview of protein expression in cells, with ultrastructural methods that offer nanometer-scale resolution. These procedures have proven successful for cells grown in a cultured monolayer, however, few solutions exist to adapt them to polarized multicellular samples.
The principal challenge in analyzing multicellular samples is identification of the Volume of Interest (VOI) and subsequent correlation of this region between light and electron microscopy images. In addition, small model organisms (including C. elegans, Drosophila, zebrafish, planaria, and tardigrades) impose additional challenges during sample preparation due to the need for proper 3D orientation of the block for sectioning. In order to recognize the structures of interest within this complex anatomy, the electron microscopist needs to have a deep familiarity with each organism`s anatomy at the ultrastructural level, which is rarely the case.
We recently developed a workflow to optimize EM sample preparation of small multicellular organisms while allowing fluorescent protein localization in ultrathin resin sections, and we have demonstrated its utility in C. elegans and Drosophila. Briefly, rapid high-pressure freezing is used to retain native fluorescence in the samples. Next, a two-step flat embedding procedure simplifies the generation of 3D maps, which are further used to orient the sample within the block and to target sectioning precisely to the VOI. This method uses a combination of anatomical cues and engineered landmarks, recognizable in both LM and EM, to orient the sample. The resulting serial sections are transferred to silicon wafer support, using a modified Array Tomography (AT) approach. The ability to retain fluorescence in the embedded sample facilitates rapid recognition of the VOI by light microscopy and serves to localize that region for subsequent SEM. Correlation within the 3D volume and reconstruction of 2D data is thus more precise and direct. These methods can be combined with recent advances in super-resolution light microscopy to accurately map protein localization to ultrastructural features. As thousands of strains expressing fluorescently-tagged proteins or cells of interest have already been generated by these research communities, this approach has the potential to be widely adopted and solve a long-standing problem in connecting traditional fluorescence microscopy experiments with ultrastructural information.
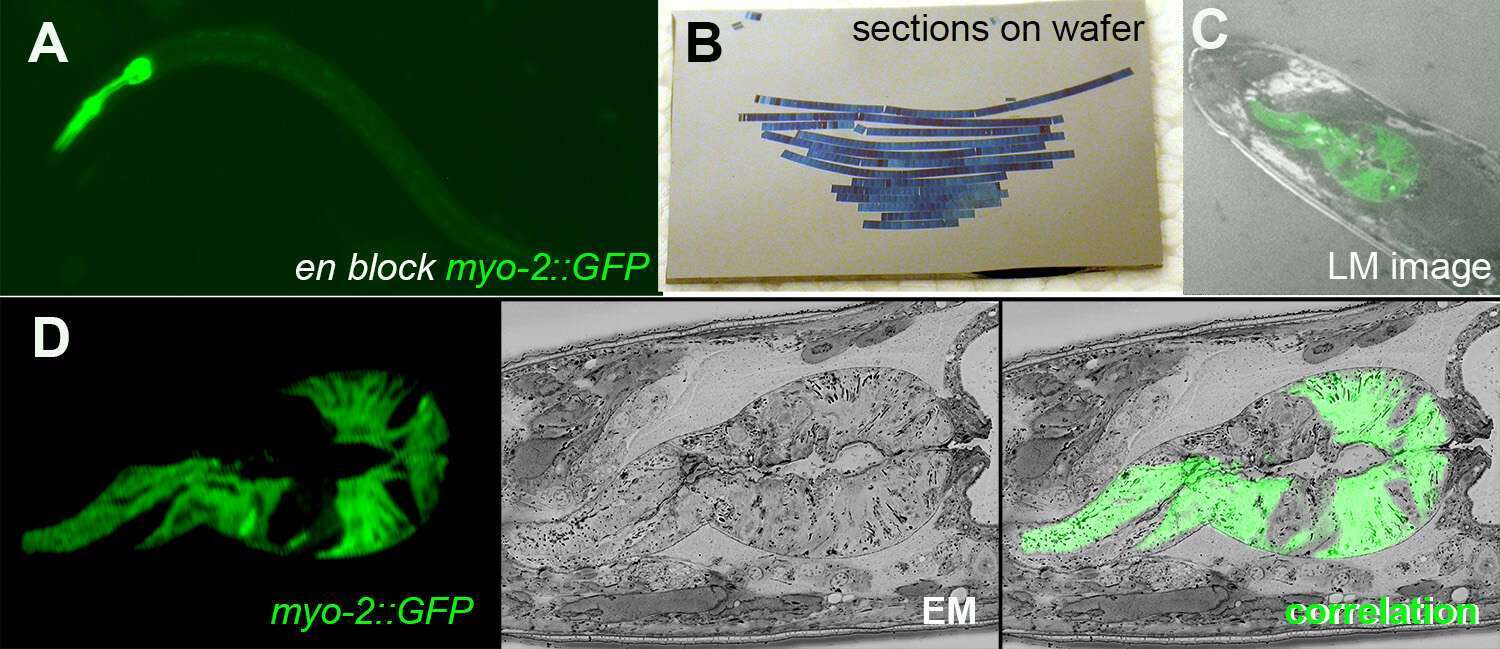
Figure 1: A. En block fluorescence of C. elegans pharyngeal muscles GFP after high pressure freezing and fast freeze substitution. B. AT sections on silicon wafer. C. A LM image of transmission and GFP fluorescence on wafer. D CLEM image of a section showing fluorescent, EM and the superposition of both.

