A TRISPYRAZOLYLBORATO IRON CYSTEINATO COMPLEX AS A FUNCTIONAL MODEL FOR THE CYSTEINE DIOXYGENASE
The cysteine dioxygenase (CDO) is a non-heme mononuclear iron enzyme that catalyzes the irreversible oxidation of cysteine by dioxygen to yield cysteine sulfinic acid, which is the first major step in cysteine catabolism in mammalian tissues.[1] While the function of many oxygenating non-heme iron enzymes could be successfully imitated within the last decades using molecular model compounds,[2] to date there are hardly any reports in the literature on biomimetic models for the CDO.
Unlike other mononuclear non-heme iron dioxygenases, the active sites of which typically contain a 2-histidine-1-carboxylate coordination motive, the iron ion of CDO is bound by three histidine ligands.[1] To this unit, the substrate cysteine coordinates through the sulfur atom and the nitrogen atom as a chelating ligand.
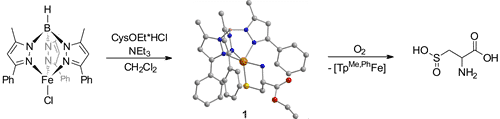
Here we present the successful coordination and dioxygenation of a protected cysteinato ligand to a (His)3FeII analogue (see Scheme):[3] In complex [TpMe,PhFeCysOEt], the TpMe,Ph ligand excellently mimics the (His)3-coordination sphere and also the function is simulated, as treatment with dioxygen mainly leads to cysteine sulfinic acid. 1 is thus the hitherto most realistic model for the active site of the CDO.
[2] a) M. Costas, M. P. Mehn, M. P. Jensen, L. Que, Jr., Chem. Rev. 2004, 104, 939-986; b) W. Nam, Acc. Chem. Res. 2007, 40, 522-531; c) I. Siewert, C. Limberg, Chem. Eur. J. 2009, 15, 10316-10328; d) S. Friedle, E. Reisner, S. J. Lippard, Chem. Soc. Rev. 2010, 39, 2768-2779.
[3] M. Sallmann, I. Siewert, L. Fohlmeister, C. Limberg, C. Knispel, Angew. Chem. Int. Ed. 2012, 51, 2234-2237.
Powered by Eventact EMS