
EXPANDING THE GENETIC CODE OF A PHOTO-AUTOTROPHIC ORGANISM
2Ilse Katz Institute for Nanoscale Science and Technology, Ben-Gurion University of the Negev, Beer-Sheva
3Niels Bohr Institute, University of Copenhagen, Copenhagen
4Mina and Everard Goodman Faculty of Life Sciences, Bar-Ilan University, Ramat-Gan
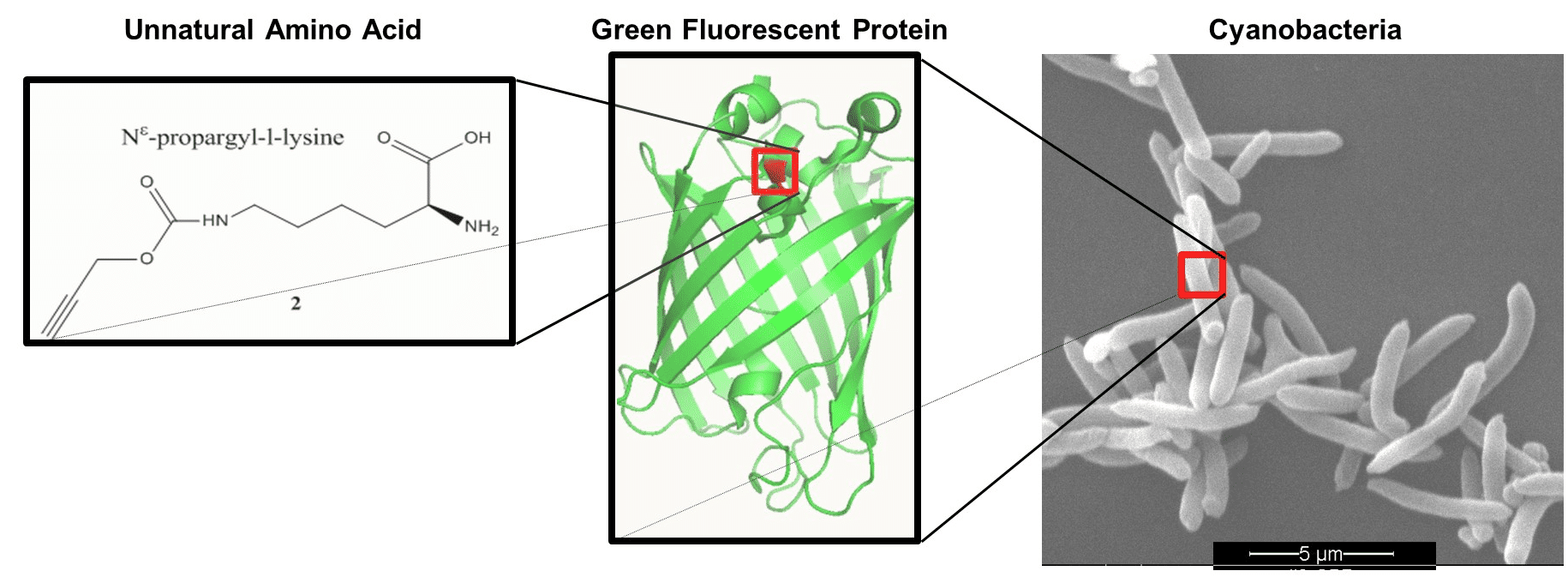
The photoautotrophic fresh water cyanobacterium S. elongatus is widely used as a chassis for biotechnology applications as well as a photosynthetic bacterial model. In this study, a method has been established to expand the genetic code of this cyanobacterium thereby enabling the incorporation of unnatural amino acids into proteins. This was achieved through UAG stop codon suppression, using archaeal pyrrolysyl orthogonal translation system. We demonstrate incorporation of unnatural amino acids into green fluorescent protein with 20±3.5% suppression efficiency (i.e. yield compared to progenitor wildtype protein). The introduced components were shown to be orthogonal to the machinery of the host. In addition, we observed that no significant growth impairment resulted from the integration of the system and no incorporation of unnatural amino acids was observed in endogenous genes that terminate with UAG. To interpret the observations we modelled and investigated the competition over the UAG codon between release factor 1 and pyl-tRNACUA. Our results suggest the existence of an unknown bacterial mechanism for discrimination against premature UAG termination by release factor 1.

Powered by Eventact EMS