
Off the Street Phasing (OTSP): Free No Hassle Haplotype Phasing for Molecular PGD Applications -AWARD NOMINEE
2Braun School of Public Health and Community Medicine, Hebrew University
Introduction: Parental phasing of mutation-flanking haplotypes is an essential, yet time-consuming, laborious, and costly pre-requisite for preimplantation genetic diagnosis (PGD) of monogenic disorders.
Aim: To validate rapid, low cost, population-assisted haplotype derivation in a pre-clinical setting.
Materials and Methods: Targeted sequencing of CFTR variants and gene-flanking polymorphic SNPs in 38 Jewish individuals from 9 different PGD families was performed at the SZMC PGD lab. Heterozygous genotype calls were both trio-phased to obtain ground-truth haplotypes, and also population-phased using Shapeit software. Reference panels for population phasing were derived from either ‘1000 Genomes’ or from 128 or 574 sample Ashkenazi Jewish whole genome sequences (kindly provided by The Ashkenazi Genome Consortium). Accuracy of resulting haplotypes was benchmarked against trio-phased haplotypes. The study population consisted of 4 subgroups of Jewish individuals: a) Non-Ashkenazi (NA; 10 negative controls); b) Full Ashkenazi (FA; 10 individuals); c) Partial Ashkenazi (13 individuals featuring at least 1 Ashkenazi and 1 non-Ashkenazi ancestor) without Ashkenazi CFTR founder mutation carriage (PANM); and d) Partial Ashkenazi (5 individuals) with W1282X CFTR Ashkenazi founder mutation carriage (PAWM).
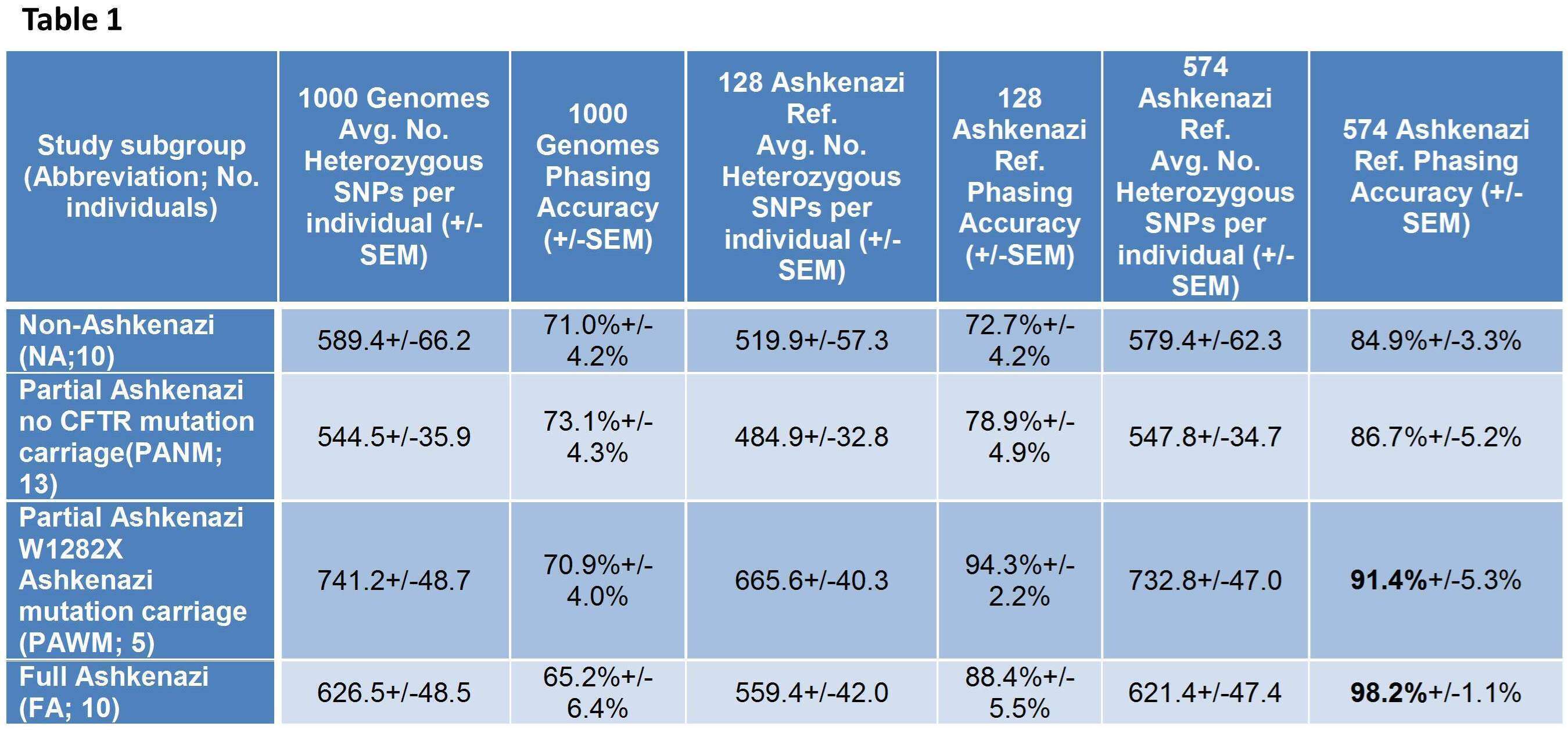
Results: Haplotype phase benchmarking results are summarized in Table 1. In general, the 574 sample Ashkenazi genome reference was the most accurate and appropriate for population-based phasing. Importantly, virtually all phase errors in the FA group (and the PAWM group, along a subregion of 3Mb) were traceable to low-coverage sequencing errors in the ground-truth.
Discussion: These striking results indicate that it may soon be possible to replace experimental haplotype phasing with clinical “OTSP,” population-based phasing, provided that one has access to an appropriate population-matched reference dataset of sufficient size.

Powered by Eventact EMS