
MOLECULARLY IMPRINTED NANOPARTICLES FOR INHIBITING RIBONUCLEASE IN REVERSE TRANSCRIPTASE POLYMERASE CHAIN REACTION
Reverse transcriptase polymerase chain reaction (RT-PCR) is considered as the gold standard in quantifying RNA and has found widespread use in food and environmental analysis as well as medical diagnostics.1 However, the accuracy of RT-PCR can be severely affected by the presence of Ribonucleases (RNases), a ubiquitous enzyme in living organisms that can degrade RNA and compromise RNA integrity.2 Molecular imprinted polymers (MIPs) which can interact with RNase at multiple binding sites including the active sites of the RNase, can be viewed as artificial inhibitors.
In this study, we proposed for the first time, to use MIPs nanoparticles (nanoMIPs) as RNase inhibitor in RT-PCR assay (Figure 1). The nanoMIPs were synthesized via a solid-phase approach and the formulation of the nanoMIPs was optimized by screening commonly used functional monomers.3 The binding capacity, binding kinetics, and the binding selectivity of nanoMIPs were investigated. The results indicate that the nanoMIPs have 1) high affinity - the dissociation constant Kd is 3.2 µg mL-1 (237nM); 2) fast binding speed – adsorption capacity saturated within 30 min; 3) good selectivity - adsorption capacity of RNase is higher than its analog. With the cavities of complementary shape, size, and chemical functional groups, the nanoMIPs could recognize and block the active sites of RNase. The results showed that the optimized nanoMIPs were able to inhibit higher than 90% of RNase, similar as the protein inhibitor. During the RT-PCR assay, the nanoMIPs managed to inhibit 10 ng of RNase in a 10 µL reaction (Figure 2). In addition, the nanoMIPs showed no interference on other enzymes like RNA reverse transcriptase and DNA polymerase. Since only a trace amount of RNase (pg - sub ng) is expected to be present in an RT-PCR reaction, the nanoMIPs could in principle replace the commercial recombinant protein inhibitors. With the competitive inhibition behavior, we believe the tailor-made nanomaterials are very promising for a variety of biological and biomedical applications.
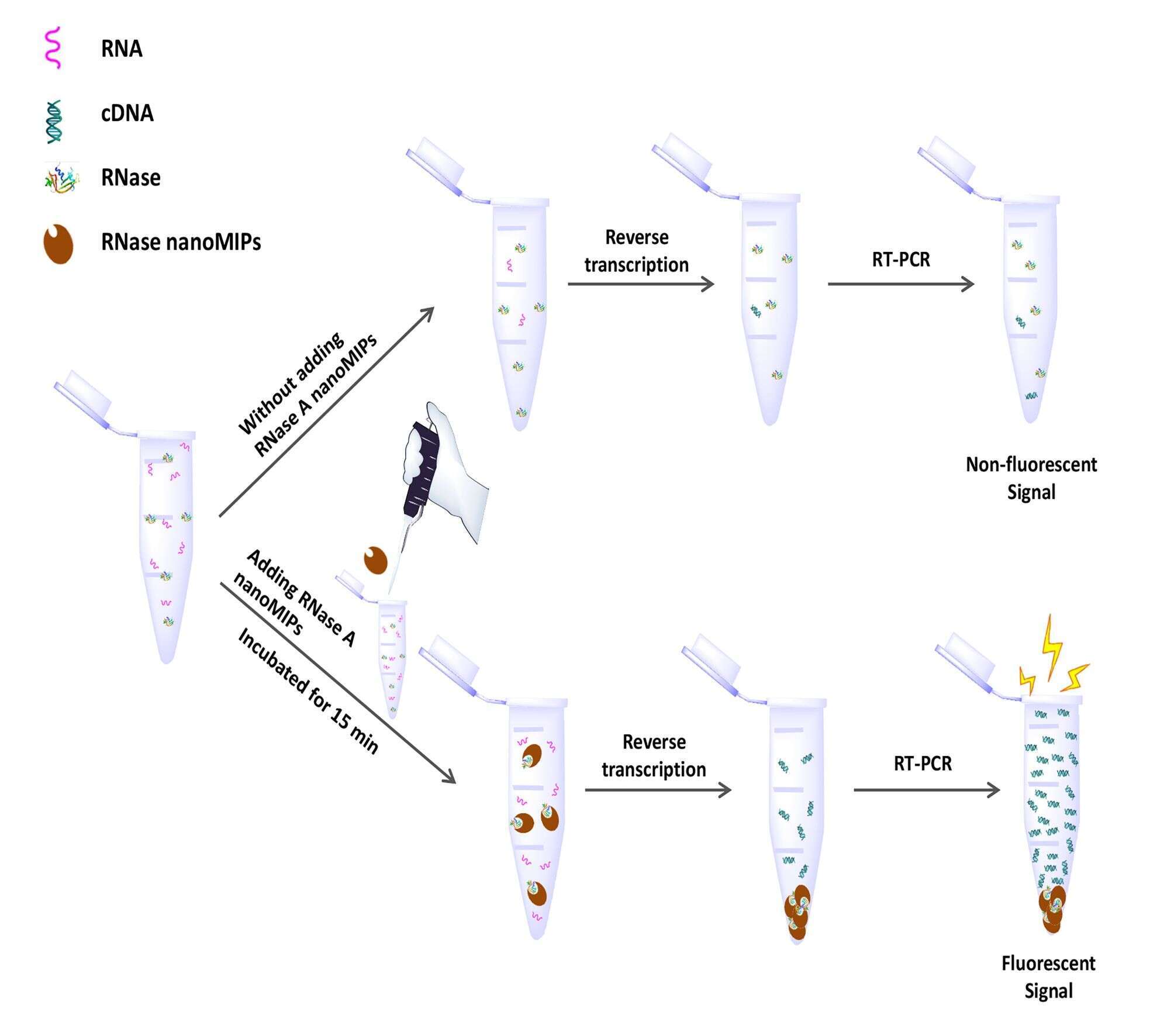
Figure 1. Schematic illustration of the application of RNase A nanoMIPs in RT-PCR assay.
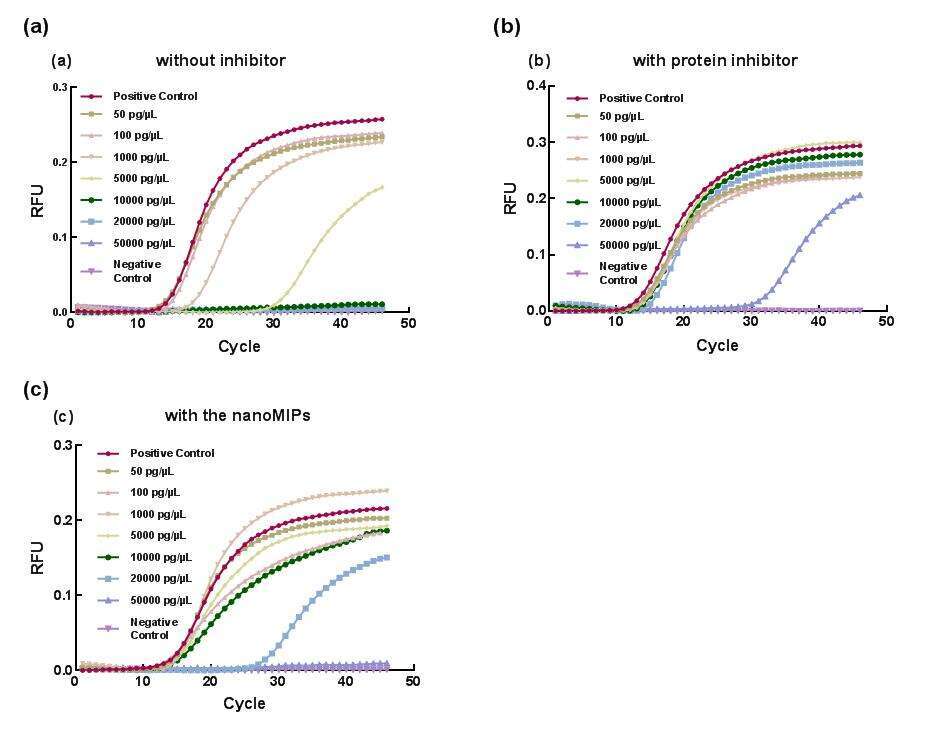
Figure 2. The real-time RT-PCR assays with (a) different concentrations of RNase A without adding inhibitors; (b) different concentrations of RNase A mixed with protein inhibitor; (c) different concentrations of RNase A mixed with nanoMIPs.
References:
(1) Tall A, Teillon A, Boisset C, Delesmont R, Touron-Bodilis A, Hervio-Heath D (2012), Real-time PCR optimization to identify environmental Vibrio spp. strains. Journal of Applied Microbiology. 113(2):361-372.
(2) Fleige S, Walf V, Huch S, Prgomet C, Sehm J, Pfaffl MW (2006). Comparison of relative mRNA quantification models and the impact of RNA integrity in quantitative real-time RT-PCR. Biotechnology Letters. 28(19):1601-1613.
(3) Canfarotta F, Poma A, Guerreiro A, Piletsky S (2016). Solid-phase synthesis of molecularly imprinted nanoparticles. Nature protocols. 11(3):443

Powered by Eventact EMS