
A SEQUENTIAL SURFACE IMPRINTING STRATEGY FOR FABRICATION OF ARTIFICIAL RECEPTORS FOR PROTEINS
Molecular imprinting is a useful method for generating artificial affinity materials with tailor-made molecular recognition sites.1-2 Due to their low cost of production and high stability, molecularly imprinted polymers (MIPs) have found wide applications in many fields. Despite rapid increase of the MIPs for protein templates, the affinity and selectivity of the polymers remain to be improved in comparison with the natural protein-based antibodies or receptors.3 Herein, we have developed a “sequential imprinting” approach which combines two different types of imprinting reactions. A significant improvement of the performance of the obtained affinity nanoparticles was demonstrated.
As illustrated in Figure 1, in the presence of the template protein, a thin MIP layer (designated as M1) was first deposited on the surface of Fe3O4@SiO2 nanoparticles by dopamine self-polymerization. Next, with the template molecules retaining in the binding sites of the obtained polydopamine, secondary functional monomers were added to precisely modify the initial binding sites in the presence of the template molecules. Different organic boronic acid molecules were employed as secondary functional monomers and their performance were investigated in detail. To prove the general applicability of the approach, two different protein templates DNase I (deoxyribonuclease I) and APE1(apurinic/apyrimidinic endonuclease/redox effector factor 1) were imprinted, respectively. The obtained artificial receptor nanoparticles were successfully used for extraction, purification and recover of the two proteins from complex biological samples.
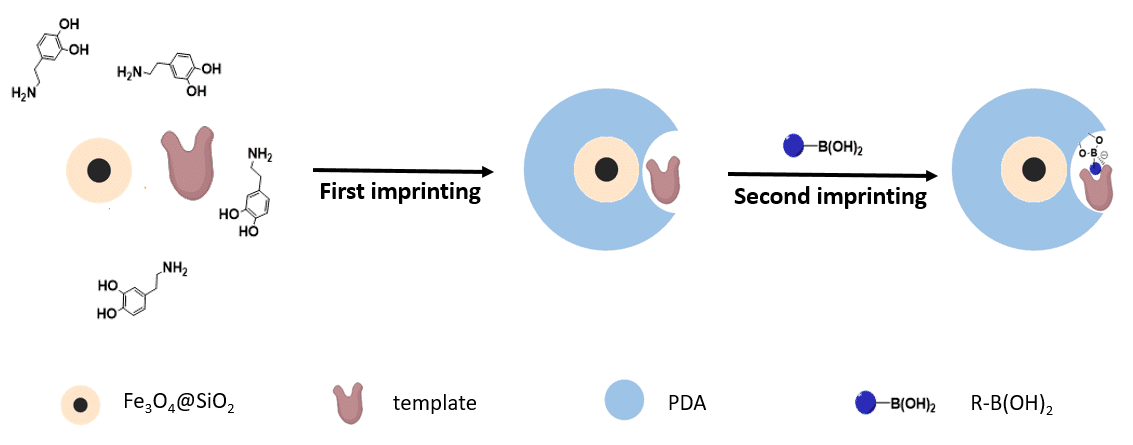
Figure 1. Schematic illustration of the synthetic approach for the sequential surface imprinting.
Acknowledgment: The work was supported by the National Natural Science Foundation of China (21775009).
References
- Haupt, K.; Mosbach, K., Molecularly imprinted polymers and their use in biomimetic sensors. Chemical reviews 2000, 100 (7), 2495-504.
- Alexander, C.; Andersson, H. S.; Andersson, L. I.; Ansell, R. J.; Kirsch, N.; Nicholls, I. A.; O`Mahony, J.; Whitcombe, M. J., Molecular imprinting science and technology: a survey of the literature for the years up to and including 2003. Journal of molecular recognition : JMR 2006, 19 (2), 106-80.
- Whitcombe, M. J.; Chianella, I.; Larcombe, L.; Piletsky, S. A.; Noble, J.; Porter, R.; Horgan, A., The rational development of molecularly imprinted polymer-based sensors for protein detection. Chemical Society reviews 2011, 40 (3), 1547-71.

Powered by Eventact EMS