In conventional microscopy, the spatial resolution is bounded by Abbe’s diffraction limit, corresponding to ~half the optical wavelength. Super resolution methods, e.g. stimulated emission depletion (STED) [1], structured illumination microscopy (SIM) [2], and localization microscopy (PALM/STORM) [3-5] have revolutionized biological imaging in the last decade, enabling the observation of cellular structures at the nanoscale. Localization microscopy relies on acquiring a sequence of diffraction-limited images, each containing point-spread functions (PSFs) produced by a sparse set of emitting fluorophores. Next, the emitters are localized with high precision. By combining the recovered emitter positions from each frame, a super-resolved image is produced with resolution typically an order of magnitude better than the diffraction limit (down to tens of nanometers).
In localization microscopy, regions with high densities of overlapping emitters pose an algorithmic challenge. This emitter-sparsity constraint leads to a long acquisition time (seconds to minutes), which limits the ability to capture fast dynamics of sub-wavelength processes within live cells. To overcome this limitation, various algorithms have been developed to handle overlapping PSFs to enable higher sample density. While successful localization of densely-spaced emitters has been demonstrated, all existing methods suffer from two fundamental drawbacks: data-processing time and sample-dependent parameter tuning. Even accelerated sparse-recovery methods such as CEL0 [6], still involve a time-consuming iterative procedure, and scale poorly with the recovered grid size. In addition, current methods rely on parameters that balance different tradeoffs in the recovery process. These need to be tuned carefully through trial and error to obtain satisfactory results; ergo, requiring user expertise and tweaking time.
Recently, we demonstrated precise, fast, parameter-free, super resolution image reconstruction by harnessing Deep-Learning [7]. Convolutional neural networks have shown impressive results in a variety of image processing and computer-vision tasks, such as single-image resolution enhancement, and segmentation. By employing a fully convolutional neural network, we reconstructed super-resolution images from very dense fields of overlapping emitters (up to 6 emitters per ). Our method, dubbed Deep-STORM, does not explicitly localize emitters. Instead, it creates a super-resolved image from the raw data directly. The net produces images with reconstruction resolution comparable or better than existing methods; furthermore, the method is extremely fast, and our software can leverage GPU computation for further enhanced speed. Moreover, Deep-STORM is parameter free, requiring no expertise from the user, and is easily implemented for any single-molecule dataset. Importantly, Deep-STORM is general and does not rely on any prior knowledge of the structure in the sample, making the method applicable to any single-molecule dataset.
We tested Deep-STORM on experimental and simulated super-resolution data, and benchmarked against the recently developed high-performance multi-emitter fitting algorithm CEL0 [6], and against the fastest multi-emitter fitting algorithm to our knowledge (FALCON [8]).
First, we reconstructed a simulated microtubule dataset available on the EPFL SMLM challenge website [9]. We quantified the quality of the results using the standard normalized mean square error: . Deep-STORM showed improved NMSE of 37% compared to 69% for CEL0 and 61% for FALCON. Second, we tested the result of Deep-STORM on experimental data obtained from Sage et al. [8], training solely on simulated data with similar experimental conditions - namely, SNR and emitter density. Deep-STORM resolves nearby lines and fine structures, and produces more continuous shapes compared to the output of CEL0 (Fig. 1) and FALCON (see SI of [7]).
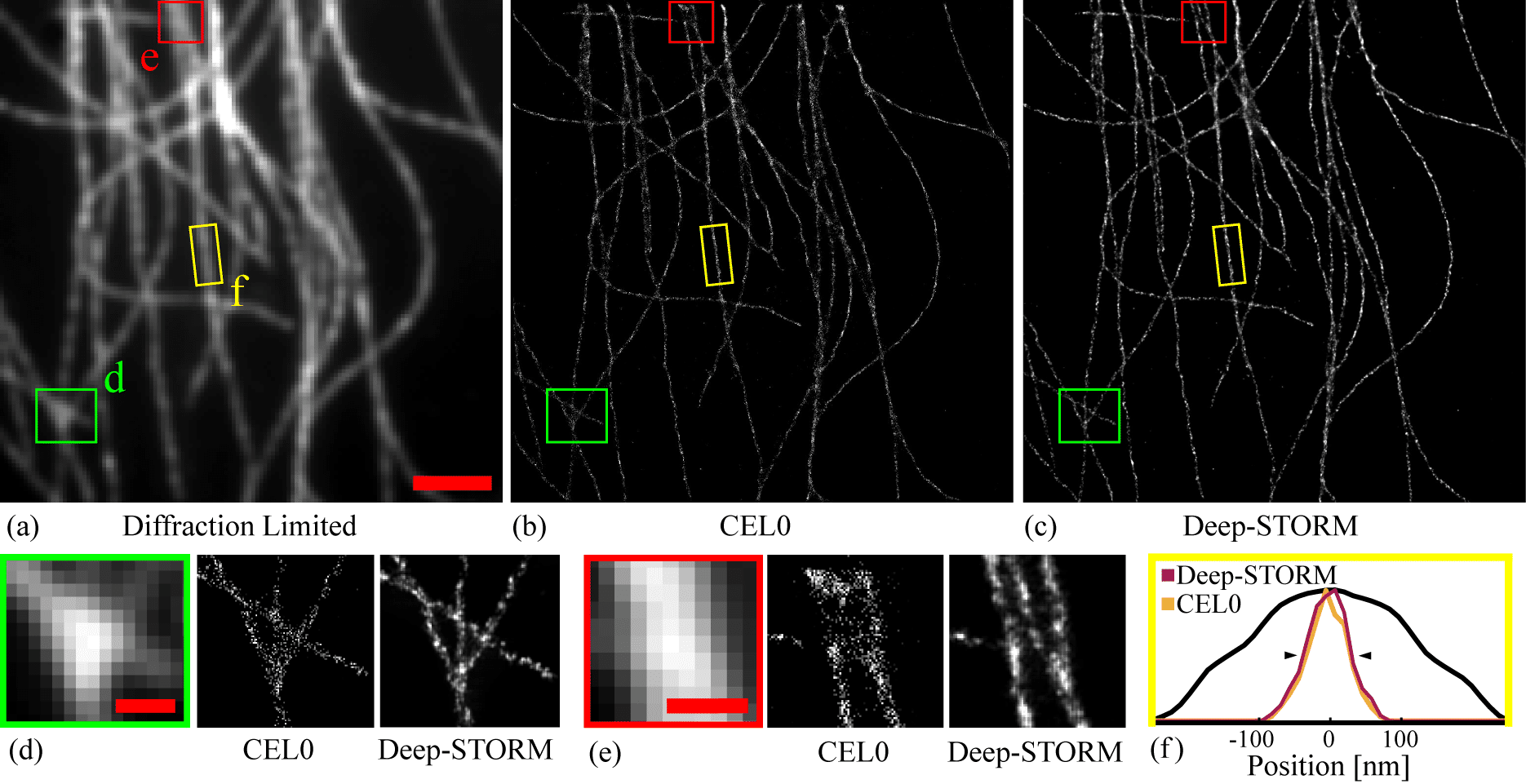
Figure 1. Experimentally measured microtubules. (a) Sum of the acquisition stack. Scale bar is 2 mm. (b) Reconstruction by the CEL0 method. (c) Reconstruction by Deep-STORM. (d)-(e) Magnified views of two selected regions. Scale bars are 0.5 mm. (f) The width projection of the highlighted yellow region. The attained FWHM (black triangles) for CEL0 was 61 nm and 67 nm for Deep-STORM. The black line shows the diffraction-limited projection.
Deep-STORM not only yields image reconstruction results that are comparable to or better than leading algorithms, but also does so ~1-3 orders of magnitude faster. Table 1 compares the run time of Deep-STORM vs. CEL0 and FALCON on both simulated and experimental microtubule datasets. Deep-STORM exhibits significantly superior runtime, especially when introducing GPU acceleration, equivalent to localizing ~20000 emitters per second, compared to ~1500 emitters per second by the fastest existing multi-emitter fitting method to our knowledge (FALCON [8]).
Table 1. Runtime comparison.
|
Dataset
|
Grid size
|
CEL0
CPU [s]
|
FALCON
CPU / GPU [s]
|
Deep-STORM
CPU/GPU [s]
|
|
Sim.
|
512 X 512
|
18677
|
1465/122
|
123/4
|
|
Exp.
|
1024 X 1024
|
175200
|
10177/434
|
715/27
|
Ultimately, the key ingredient in training neural networks is the training set. The best training set should include the aberrations in the experimental imaging system; however, very large data sets are typically used to train a deep neural network, and obtaining massive amounts of experimental images is not straight-forward. However, we found that a reasonable number of experimental images (100) is sufficient to train a high-quality net. We trained and tested Deep-STORM on a sample containing randomly scattered fluorescent quantum dots to evaluate the performance on experimental data with a variety of SNR conditions encountered in single-molecule data sets, and at high density. Comparing to reconstruction of the same images using a net trained on simulated data, we found that the experimentally-trained net outperforms the simulated net, detecting 96% compared to 88% of the emitters, with a reduced false positive rate of 1.6% compared to 8.7%. This test demonstrates that while simulated data can serve as excellent training data - experimentally obtained images are even better. Additionally, a high-quality reconstruction net can be trained using a small number of experimentally measured images.
To conclude, Deep-STORM combines state-of-the-art resolution enhancement, unprecedented speed, and high flexibility (parameter-free operation). This combination produces a technique potentially capable of video-rate analysis of super-resolution localization-microscopy data that requires no expertise from the end user, overcoming some of the most significant limitations of existing localization methods.
References:
[1] S. W. Hell and J. Wichmann, “Breaking the diffraction resolution limit by stimulated emission: stimulated-emission depletion fluorescence microscopy,” Opt. Lett., vol. 19, pp. 780–782, Jun 1994.
[2] M. Gustafsson, “Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy.” J. microscopy 198, 82–7 (2000).
[3] E. Betzig, et al., “Imaging intracellular fluorescent proteins at nanometer resolution,” Science (2006).
[4] S. T. Hess, T. P. Girirajan, and M. D. Mason, “Ultra-high resolution imaging by fluorescence photoactivation localization microscopy,” Biophys. J. 91, 4258 – 4272 (2006).
[5] M. J. Rust, M. Bates, and X. Zhuang, “Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM),” Nat. Methods 3, 793–795 (2006).
[6] S. Gazagnes, E. Soubies, and L. Blanc-Féraud, “High density molecule localization for super-resolution microscopy using CEL0 based sparse approximation,” ISBI 2017-IEEE Int. Symp. on Biomed. Imaging p. 4 (2017).
[7] E. Nehme, et al., “Deep-STORM: Super resolution single molecule microscopy by deep learning“, arXiv:1801.09631. Accepted to Optica (2018).
[8] J. Min, et al., “FALCON: fast and unbiased reconstruction of high-density superresolution microscopy data,” Sci. Reports 4, 4577 (2015).
[9] D. Sage, et al., “Quantitative evaluation of software packages for single-molecule localization microscopy,” Nat. Methods 12, 717–724 (2015).