Rationale: The cardiomyocyte is a long living cell that is dedicated to operating and maintaining a macromolecular contractile machine - the sarcomere. Sarcomeric proteins have limited half-lives, therefore they require continuous replacement. Such maintenance is challenging since transcription and translation are ‘noisy’ biological processes, and cardiomyocytes need to maintain all their sarcomeres simultaneously while preserving their stoichiometry in order for normal contractile activity to proceed.
Objective: Our study aims to delineate a working model of sarcomeric homeostasis, addressing the current gap in knowledge of the molecular mechanisms used by cardiomyocytes to efficiently maintain all their sarcomeres simultaneously.
Methods and Results: We used single-molecule fluorescence in-situ hybridization (smFISH) to visualize mRNA, and demonstrated that sarcomeric mRNA localizes to the sarcomere (Figure 1). By using MATLAB analysis of smFISH images, we were able to analyze transcriptional dynamics and showed that transcription rate and mRNA content is variable in cardiomyocytes. Transcriptional variability was validated via single-cell RT-qPCR (real-time, quantitative polymerase chain reaction). Protein synthesis was visualized by OPP (O-propargyl-puromycin) assay, both in cultured cells and in–vivo, which demonstrated localized protein synthesis at the sarcomere (Figure 2). In-vivo OPP assay was performed by echo-quided intramyocardial injection of OPP and imaging of cardiac sections. Using automated analysis with Cellprofiler, we show that protein synthesis rates vary widely between cells, yet protein content remains relatively constant. Unincorporated sarcomeric proteins are rapidly degraded by the localized ubiquitin-proteasome system, offsetting the transcriptional and translational variability (Figure 3).
Conclusions: Three distinct mechanisms are responsible for the maintenance of the sarcomere: mRNAs encoding for sarcomeric proteins are localized to the sarcomere, ribosomes are localized to the sarcomere with localized sarcomeric protein translation, and finally, a localized E3 ubiquitin ligase and localized ubiquitination allow efficient degradation of excess unincorporated sarcomeric proteins. We show that these mechanisms are distinct, required, and work in unison, to ensure both spatial localization, and to overcome the large variability in transcription of sarcomeric genes. Cardiomyocytes simultaneously maintain all their sarcomeres using localized translation-degradation ‘facilities’ where proteins are continuously and locally synthesized at high rates, and excess proteins are continuously and locally degraded, providing a very robust yet adaptable system.
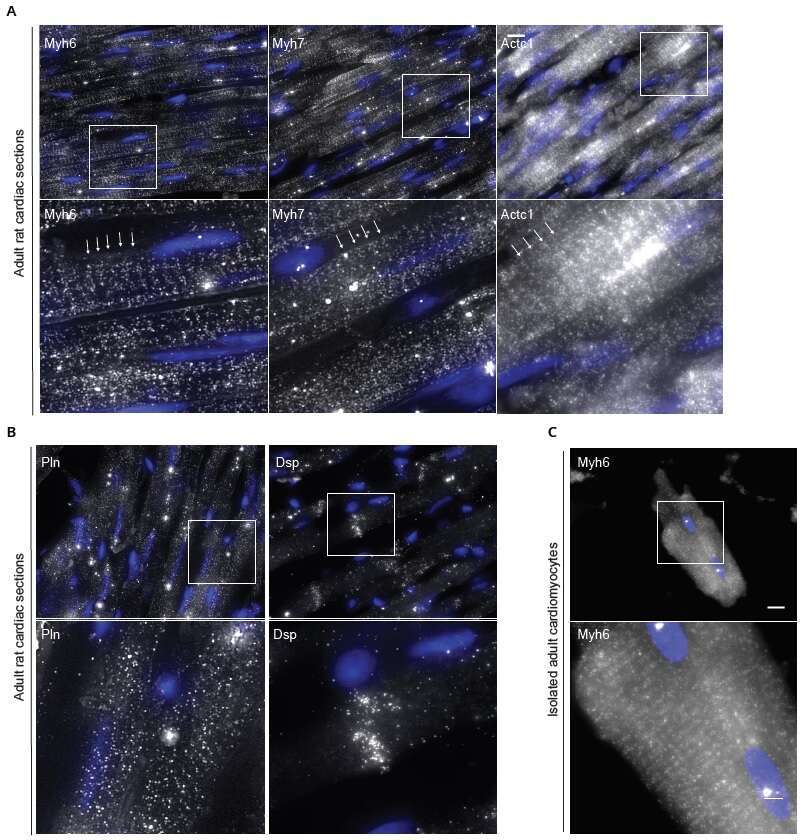
Figure 1 Sarcomeric mRNA transcripts are localized in cardiomyocytes.
(A) Adult rat cardiac tissue sections. Sarcomeric mRNA transcripts (myh6, myh7, actc1) are localized to the sarcomere. Sarcomeres indicated by arrow.
(B) Non sarcomeric genes show a different pattern - Pln appears diffuse; Dsp localizes to the intercalated disc.
(C) Sarcomeric localization of myh6 mRNA in isolated adult cardiomyocyte.
smFISH signal (white), DAPI (blue). Scale bar, 10µm; inset, 3µm.
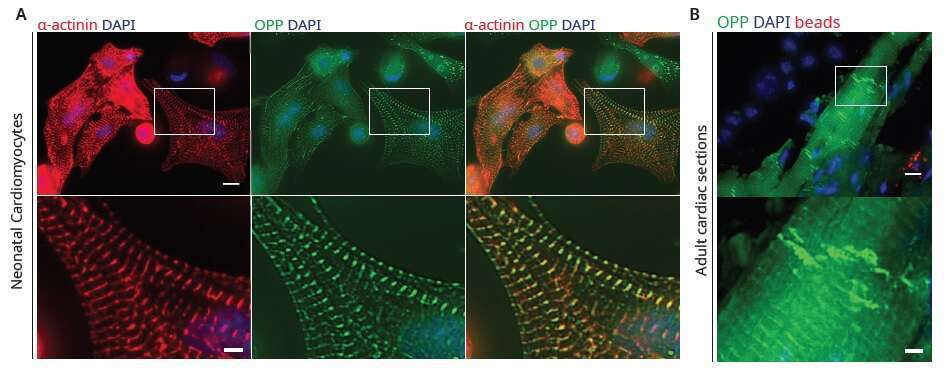
Figure 2 Sarcomeric localization of protein translation in cardiomyocytes.
(A) Imaging of newly synthesized proteins by pulse labeling neonatal rat cardiomyocytes with OPP for 30 min (green), shows that translation is highly localized to the sarcomere. Cells were co-immunostained with sarcomeric α-actinin antibodies (red), and DAPI (blue). Scale bar, 10µm; inset, 3µm.
(B) In vivo imaging of newly synthesized proteins with OPP (green) shows high localized translation in sarcomeres and at the intercalated discs (arrow). Fluorescent microbeads (red) mixed with the OPP mark the injection site. Scale bar, 10µm; inset, 3µm.
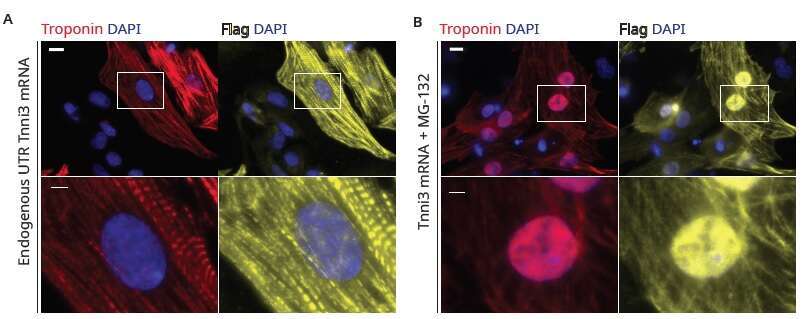
Figure 3 Neonatal rat cardiomyocytes transfected with TnnI3:Flag modified mRNA.
(A) Under regular culture conditions, normal cardiomyocyte and sarcomere structure. Overexpression of troponin does not lead to sarcomere derangement.
(B) Inhibition of proteasome-mediated degradation with MG-132 leads to nuclear accumulation of overexpressed troponin.
Red - Troponin; Yellow - Flag; Blue - DAPI. Scale bar, 10µm.