Sometimes new is the well forgotten old! Sophisticated and unique high resolution methods can be found across literature. Yet, not all studies require the finest resolution. Here, we explore the combination of good "old fashion" techniques like cryostat thick sectioning and confocal as well as "new generation" light microscopy based 3D method.
The correlative microscopy methods are presented at three biological levels: whole organ, tissue and intracellular. For the whole organ we use HREM, High-Resolution Episcopic Microscopy (developed by Tim Mohun, CR-UK). Using HREM we can visualize embryos, plants and small organs. Its resolution allows the identification of individual nuclei, nerves and blood vessels, which may not be detectable using lower-resolution techniques such as optical projection tomography (OPT), micro-MRI or micro-CT. It allows histological staining that can be segmented and quantified in 3D.
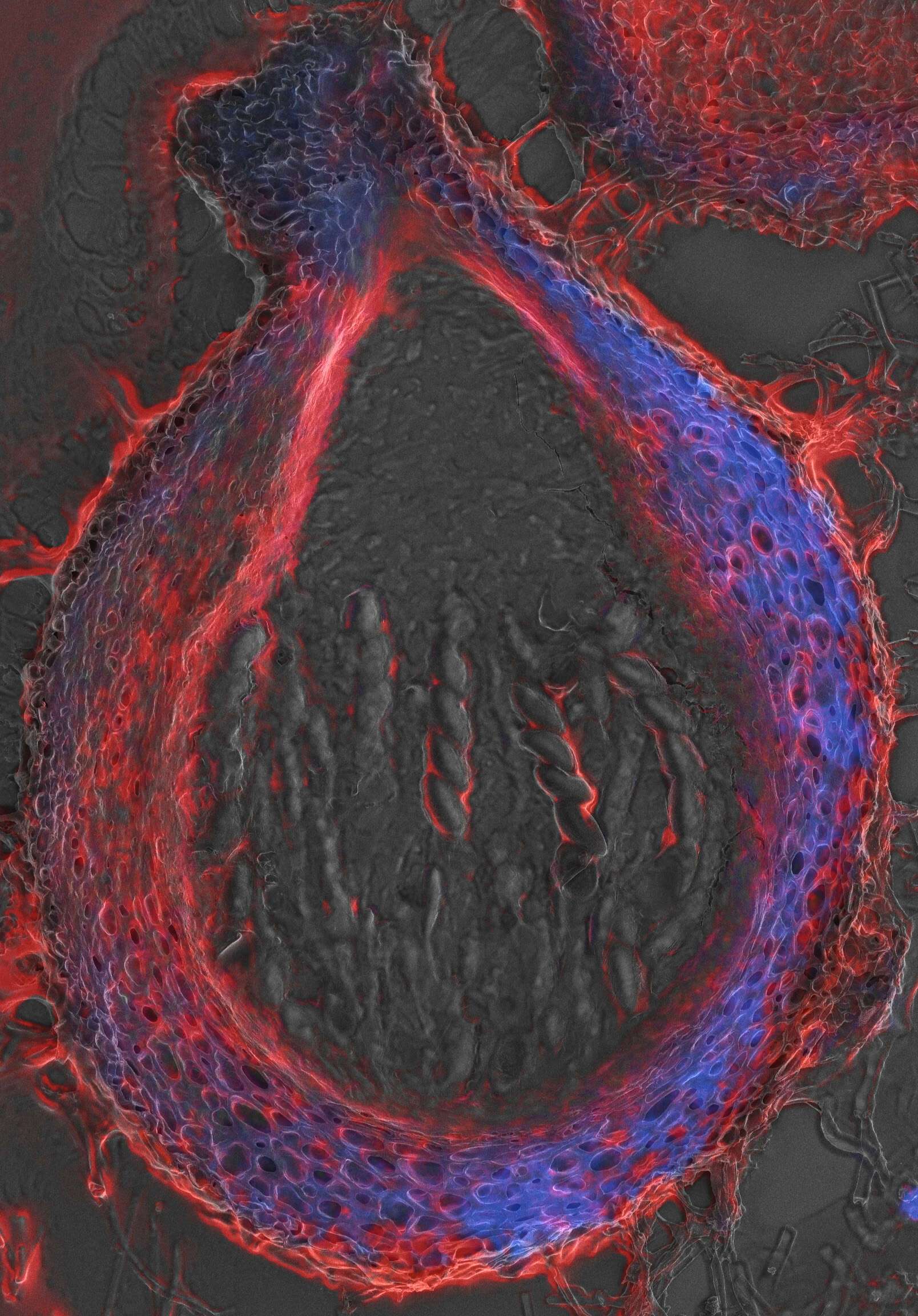
For the tissue level we used a combination of thick 10-20 micron cryostat sections either pre- or post-stained with fluorescent markers. Following dry/wet fluorescence imaging samples were further prepared for SEM. This combination allows spatial assessment of fluorescent markers in the larger context of the tissue in EM resolution. Looking at tissues such as Drosophila reproductive organs, Fruit cuticle & cell wall and Fungal sexual fruiting bodies indicated a very friendly and versatile method.
For the intracellular level, fluorescent in situ hybridization (FISH) with specific DNA probes in somatic and meiotic cereal anther tissues were used. Following confocal imaging samples were dehydrated and prepared for corresponding HRSEM via XY specific coordinates. This method proved useful for the study of nuclear and chromosomal patterns of tandem repeats and loose-end DNA fragments.