
Predicting Cancer Resistance Genotypes
It is inspiring that while we still barley understand how to identify, attack and destroy cancer cells and prevent it from development, there are many species already “solved” this. Surprisingly it is clear that the solution for cancer exists, and more important it probably not so complex where many species independently evolved the “anti -cancer secret sauce”. For example, some species live underground with high oxidative stress, which is known to enhance cancer. Instead, some of them evolved to be cancer resistant. In addition, the big animals like whales and elephants should have very high cancer rates compared to the smaller animals due to their larger number of cells. However, their cancer rates are much lower. This means that they developed cancer resistance mechanisms.
For years, the focus has been to investigate the genetics of cancer susceptibility, as a response to the suffering that cancer causes. Meanwhile, the genetics of cancer resistance have remained largely uninvestigated. However, “an ounce of prevention is worth a pound of cure”. Our ultimate goal is to develop cancer vaccine. Our vaccine will be given to the newborns and immunize their bodies against cancer simply as the known regular vaccinations immunize our bodies against possible infections. To develop our cancer vaccine we need to understand exactly how organisms get immune for cancer and try to mimic their resistance mechanisms in human. In other words, we are cheating from the billions of years of experience of resistant species against cancerous stresses.
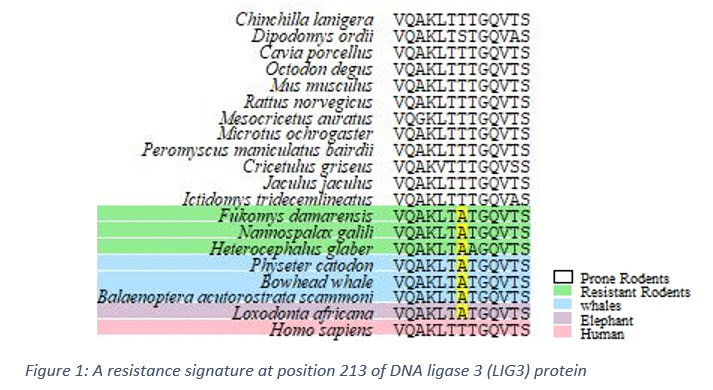
At this stage of our research, we utilized comparative genomics approaches to detect the variants and regions of the DNA that led to the resistance mechanisms. More precisely, we generated multiple sequence alignments of the whole proteomes of several mammals. These mammals were divided into two groups; cancer resistant and cancer prone. We developed a score that takes into account amino acids properties and entropy of each column in each alignment to select positions that showed significant variations of this score between the resistant and the prone groups as shown in the figure below. We found many such signatures that is located in several active domains of cancer suppressor genes. To test these signatures we are preparing CRISPER experiments on cells to investigate the effect of these signatures on its cancer resistivity.
Tel: 972-3-6384444 Fax: 972-3-6384455
cancerconf@ortra.com

Powered by Eventact EMS