
Contributed
Deciphering the architecture of SMC protein complexes
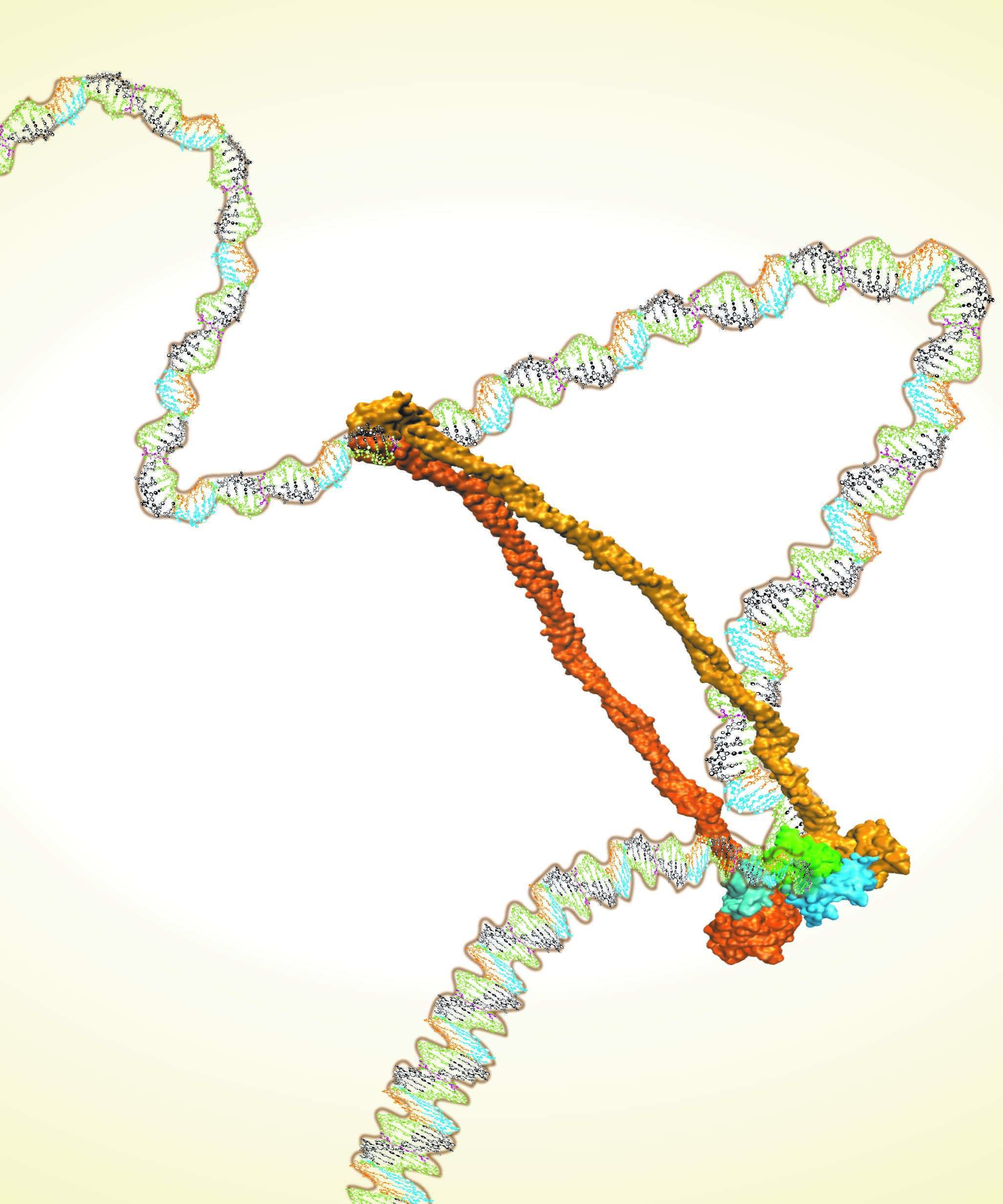
Understanding how chromatin is organized within the nucleus and how this 3D architecture influences gene regulation, cell fate decisions and evolution are major scientific questions. Despite recent progress, little is known about the mechanisms underlying chromatin structure and how it can be established, reset and maintained. One prominent hypothesis suggests that loop-extrusion motors dynamically bind and translocate segments of DNA to form structural loops. These motors are commonly proposed to be Structural Maintenance of Chromosomes (SMC) protein complexes. While limited structural data exists for the proteins that comprise the SMC complexes, the complete structure of the entire complex remains unknown.
Using an integrative approach combining both crystallographic data and co-evolutionary information, we predict an atomic-scale structure of the whole condensin complex and settle the controversy regarding the stoichiometry of the complex [1]. Using molecular-dynamics simulations, our analysis also reveals several additional configurational states of several SMC complexes. These are likely involved with the functional opening and closing of these rings, and their various interactions with chromosomes. This work sheds light on the molecular mechanisms behind the establishment and the arrangements of the 3D genome.
- D. Krepel, R.R. Cheng, M. Di Pierro and J.N. Onuchic “Deciphering the Structure of The Condensin Protein Complex”, Proc. Natl. Acad. Sci USA, in press, doi.org/10.1073/pnas.1812770115 (2018).
Powered by Eventact EMS