
Flash
Investigating the insulin effect on the α-synuclein aggregation in Parkinson`s disease
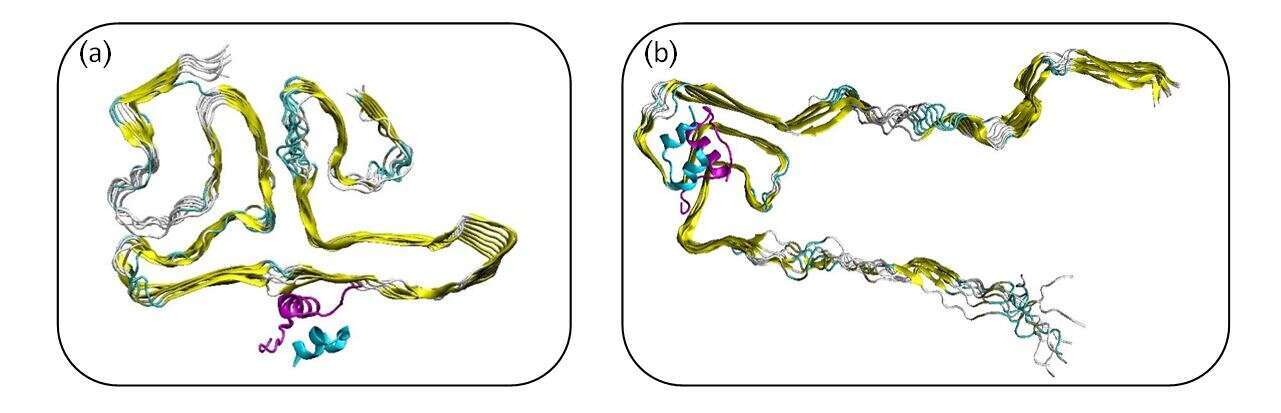
Parkinson`s disease (PD) is characterized by Lewy bodies which their major components are α-synuclein (AS) aggregates. Insulin accesses the cerebrospinal fluid cross the blood brain barrier and regulates brain function. Current and future pharmacological strategies being developed to restore neuronal insulin signaling as a potential strategy for slowing PD. It is known that the “non-amyloid β component” (NAC) domain in AS plays role in AS aggregation. The study of the self-assembly of the NAC domains is crucial in order to understand the molecular mechanisms of AS aggregation. Recently, we presented polymorphic structures of the NAC fibrils (M. Pollock-Gagolashvili, Y Miller, ACS Chem. Neurosci. 8, 2613 (2017)) and polymorphic structures of full-length AS fibrils. We then predicted twelve possible insulin-NAC fibrils models, in which the interactions between insulin and NAC fibrils are based on homology considerations, i.e., identities and similarities between the amino acids. The current study examines the effect of insulin on polymorphic full-length AS fibrils (Figure 1). Using molecular dynamics simulations, we first found that insulin can be attached to the NAC fibrils and the interactions are strong and stable along all the time scale of the simulation and can bind in various domains in NAC fibrils. We found that insulin does not affect the secondary structure of the NAC fibrils. We further examined the interactions between insulin and full-length AS fibrils and found that insulin does have an effect on its secondary structure.
Figure 1: Polymorphic Insulin-AS constructed models. (a) based on Bloch & Miller model (D. N. Bloch, Y Miller ACS Omega 2, 3363 (2017) (b) based on Rienstra`s group (Tuttle, M. D. et al. Nat. Struct. Mol. Biol. 23, 409 (2016)).
Powered by Eventact EMS