
Towards catalytic antibiotics: Redesign of aminoglycosides to catalytically disable bacterial ribosomes
2Faculty of Pharmacy with the Laboratory Medicine Division, Medical University of Warsaw, Warsaw, Poland
Aminoglycosides are antibiotics that selectively target the bacterial ribosome leading to a series of miscoding events, which result in bacterial death. Unfortunately, the prolonged clinical use of antibiotics has resulted in rapid evolvement of resistant bacterial strains that severely restrict their use. This public health concern has revived an interest in the discovery and development of novel strategies that can address the problem of growing antibacterial resistance. We set out to explore the potential of catalytic antibiotics as a new paradigm in antibiotics research. Nowadays such antibiotics do not exist and this new approach can lead to a new era of antibiotics development.
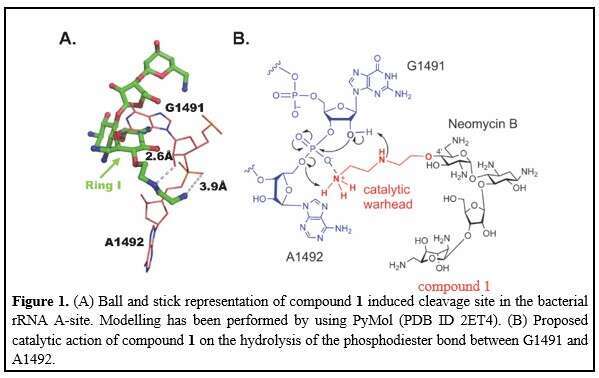
Using high resolution structures of aminoglycosides bound to their ribosomal target, along with molecular dynamics simulations, we were able to rationally design a series of structures with a potential to specifically bind and catalytically cleave a highly conserved region within the ribosomal decoding site RNA; therefore, resulting in a fast and irreversible inactivation of translation machinery. The designer structures contain as a scaffold the natural aminoglycoside antibiotics neomycin B and kanamycin B, substituted with di-amine warhead at 4` position. In attempts to enhance catalytic efficiency, kanamycin B scaffold was also bis-substituted at 3’ and 4’ positions. The new mono-substituted derivatives showed significant antibacterial activity against wild-type bacteria and were especially potent against resistant and pathogenic strains including Pseudomonas aeruginosa and Methicillin-resistant Staphylococcus aureus (MRSA).
The design principles along with the synthesis and preliminary biological evaluation of the target structures will discussed.
Powered by Eventact EMS