
Duplicate sample analysis of the short fraction of cell-free DNA identifies circulating tumor DNA in glioblastoma patients
2Huntsman Cancer Institute, University of Utah, Salt Lake City, UT, USA
3Department of Pathology, University of Utah, Salt Lake City, UT, USA
4ARUP Laboratories, ARUP, Salt Lake City, UT, USA
5Department of Neurological Surgery, University of Utah, Salt Lake City, UT, USA
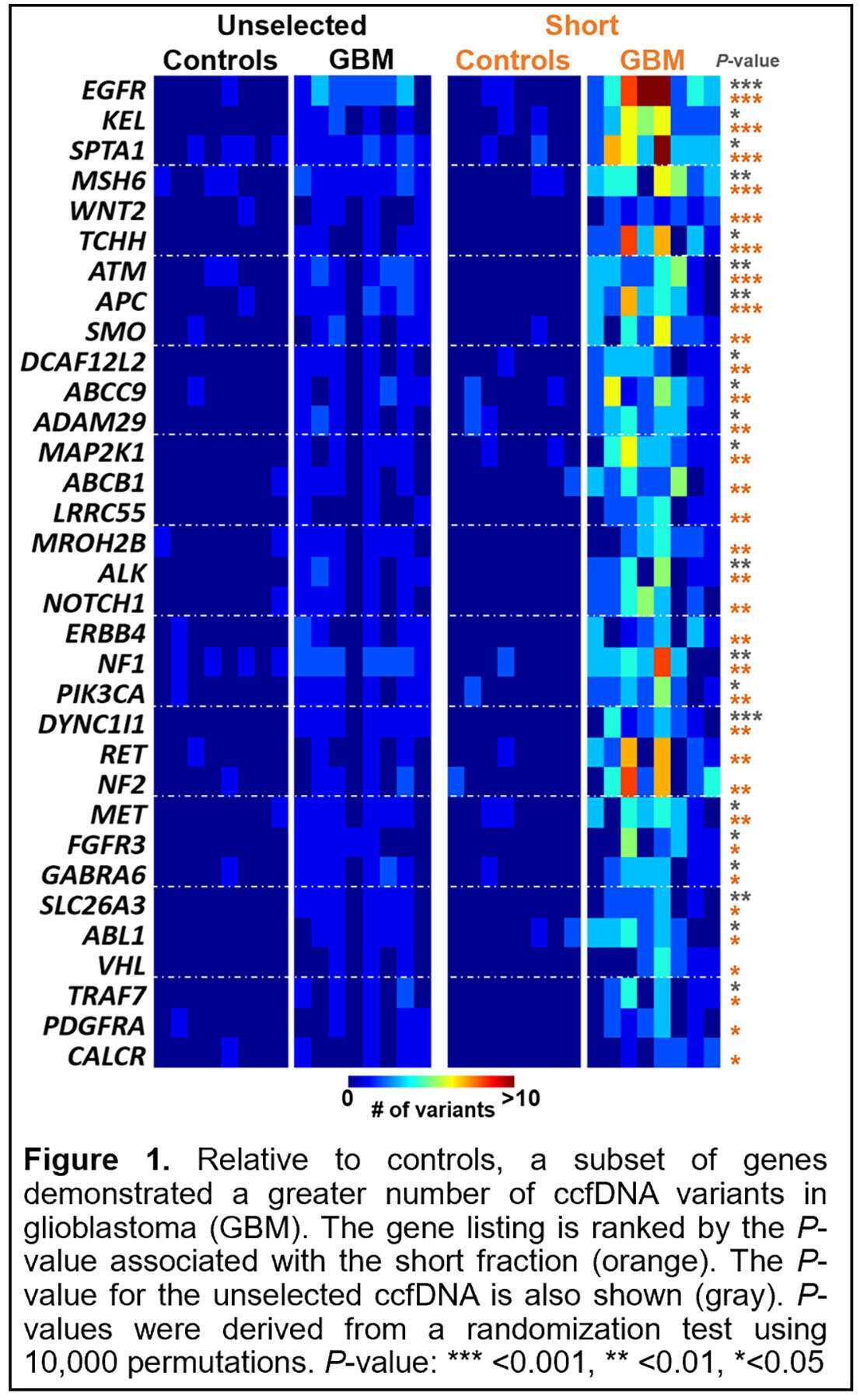
Detection of glioblastoma-derived circulating cell-free DNA (ccfDNA) has proven challenging. Glioblastoma’s vast intra-tumor genetic heterogeneity has limited use of molecular profiling from a focal tissue sample to guide circulating tumor DNA (ctDNA) detection. Here, we sought to detect glioblastoma-derived ctDNA by employing methodologies to enhance ctDNA sensitivity and specificity. Specifically, isolation of shorter ccfDNA fragments relative to ccfDNA’s principal mononucleosomal peak (<150 bp vs. 167 bp, respectively) can be used to enrich for ctDNA and reduce false positives associated with next-generation sequencing (NGS). Moreover, recent data supports using sample duplicates to further suppress NGS-related noise to search for ctDNA without a priori knowledge of somatic tumor variants. The ccfDNA from eight healthy controls and eight glioblastoma patients was isolated and all subsequent steps were performed in duplicate including isolation of the short ccfDNA fraction (<150 bp). Samples were capture-enriched using a custom-designed, glioblastoma-targeted NGS panel (118 genes, 124 kb). Non-reference alleles (NRAs) in exon consensus sequences present in duplicate were tabulated. In the short ccfDNA fraction, there were significantly more NRAs in glioblastoma patients (i.e., potential tumor-derived variants) compared to healthy controls (144.1±98.2 vs. 12.1±5.5 NRAs, respectively; P<0.001). In glioblastoma patients, there were significantly more NRAs in the short ccfDNA fraction compared to the original unselected ccfDNA sample (144±1 vs. 38.7±19.3 NRAs, respectively; P<0.011). A subset of genes within glioblastoma samples showed increased variant frequency compared to healthy controls, which was more prominent in the short fraction (Figure 1). Selection for short ccfDNA fragments coupled with duplicate analysis detects glioblastoma-derived ctDNA through concomitant improvements in both sensitivity and specificity during untargeted searches employing panel-based NGS.
Powered by Eventact EMS