
Genetic code expansion of Pseudomonas aeruginosa and bio-tracking of flagella filaments
2Department of Biotechnology Engineering, Ben-Gurion University of the Negev, Beer-Sheva, Israel
3Department of Chemistry, Ben-Gurion University of the Negev, Beer-Sheva, Israel
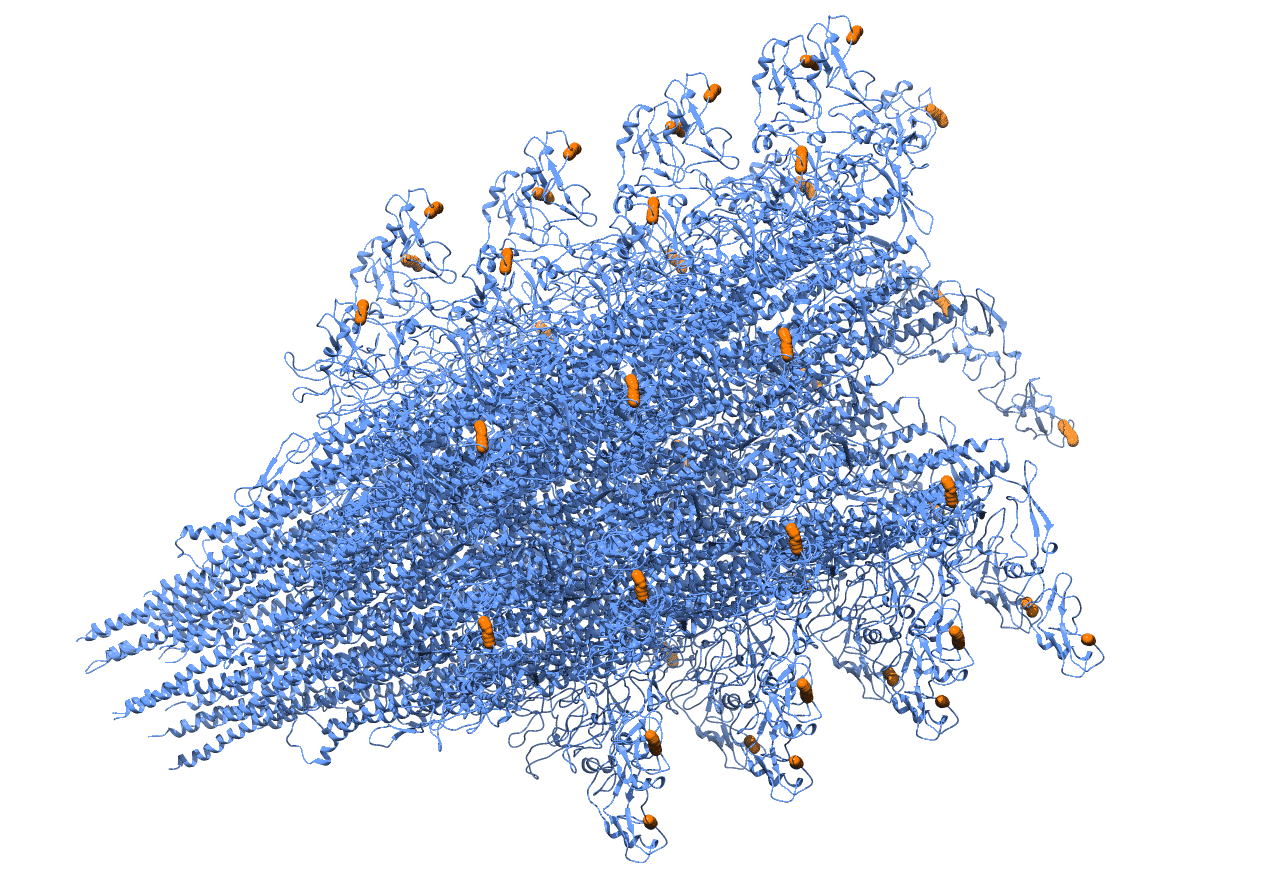
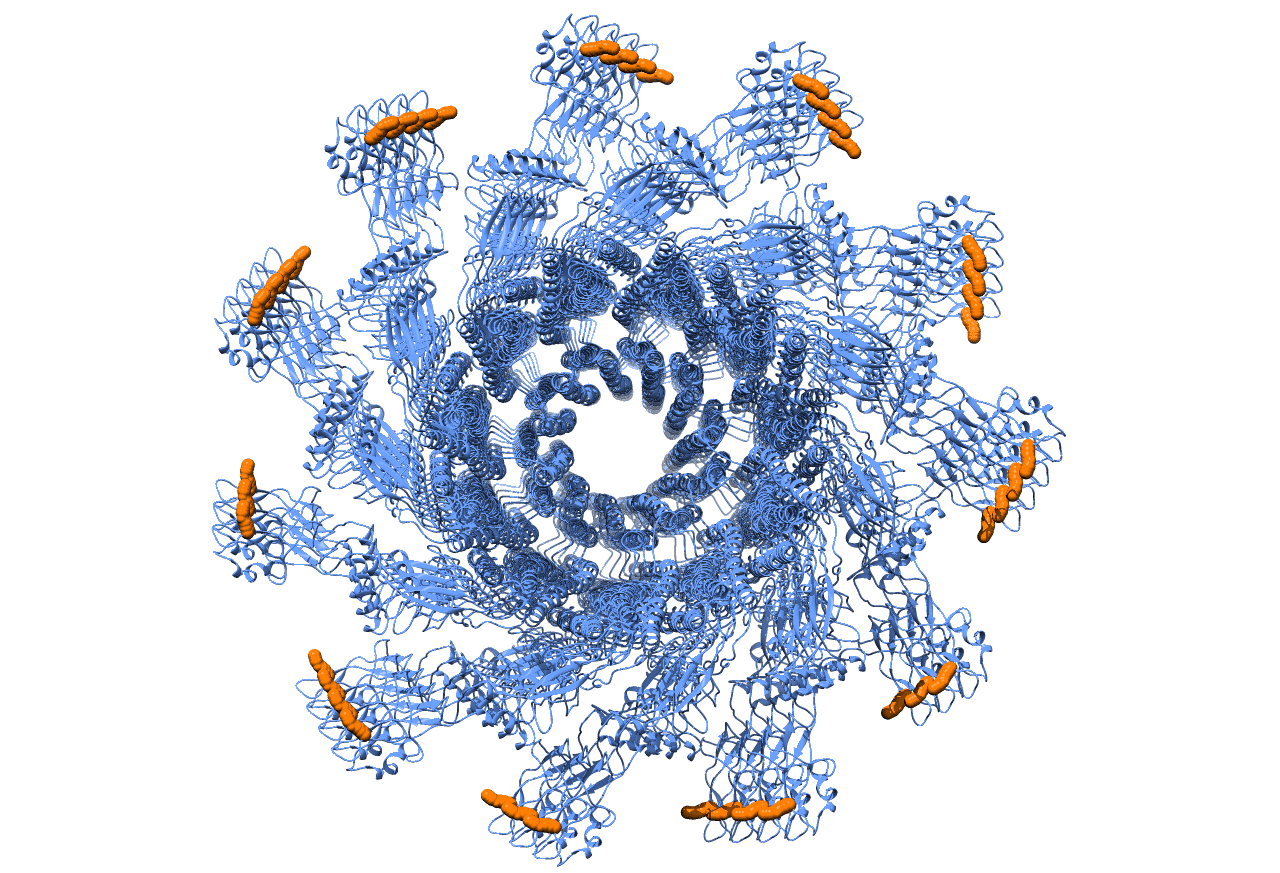
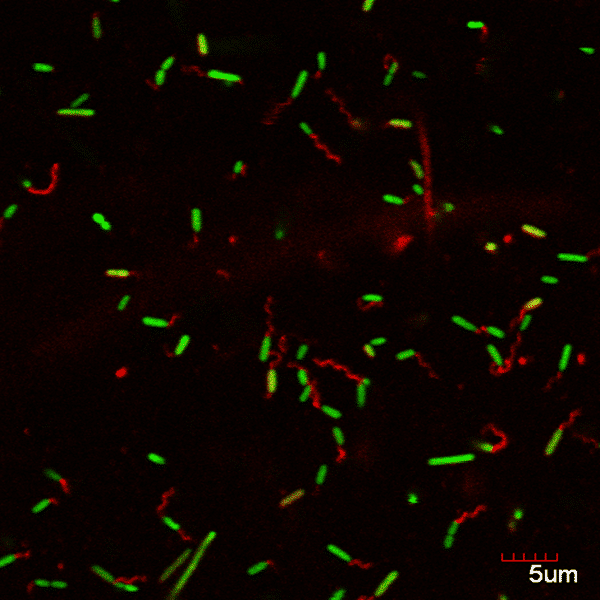
Genetic code expansion, the technology for incorporating unnatural amino acids into proteins, serves as a robust tool for protein engineering in different organisms. Here, demonstrated and employed for the first time, is the incorporation of an unnatural amino acid into the well-studied bacterium, Pseudomonas aeruginosa. By incorporating an unnatural amino acid into Pseudomonas aeruginosa flagellin, the monomer composing the bacterial flagella, a microstructure with functional anchor points was created (Fig 1a, 1b). Fluorescence imaging verified the existence of the desired microstructure (Fig2). Next, the labeled microstructure was tracked during biofilm generation and maturation. The establishment of the genetic code expansion technology elucidates new knowledge regarding biofilm maturation mechanism and flagella dynamics and allows further manipulation of Pseudomonas aeruginosa proteins. Our long-term goal in that respect is to utilize our genetically expanded Pseudomonas aeruginosa as a robust tool box for studies of quorum sensing mechanisms in bacteria.
Powered by Eventact EMS