
ForSDAT: A fully automated data analysis toolkit for SMFS measurements
Single molecule force spectroscopy (SMFS) using atomic force microscopy (AFM) is a frequently used to study ligand-receptor interactions, adhesion events and protein folding. The common setup of such an experiment includes the approach of a functionalized AFM probe to the substrate of interest. These two entities interact when the probe approaches the surface and when the probe is retracted the pull-off force can be recorded. SMFS experiments typically produce thousands of force vs. distance curves. The manual analysis of these data is cumbersome and prone to human error and bias, and is usually very time consuming. Most available data analysis software offer limited automation of specific interaction detection. Moreover, the automatic open-source programs commonly employ force thresholding to distinguish specific interactions from non-specific interactions. This approach is sensitive to low signal/noise ratio (SNR) and requires optimization for each set of results.
Here, we present ForSDAT, the Force Spectroscopy Data Analysis Toolkit, as a tool for completely automated data analysis. Our results show the high success rate of the algorithms developed for ForSDAT. Moreover, ForSDAT is a modular toolkit which can be easily fit to different analysis processes and extended with custom components.
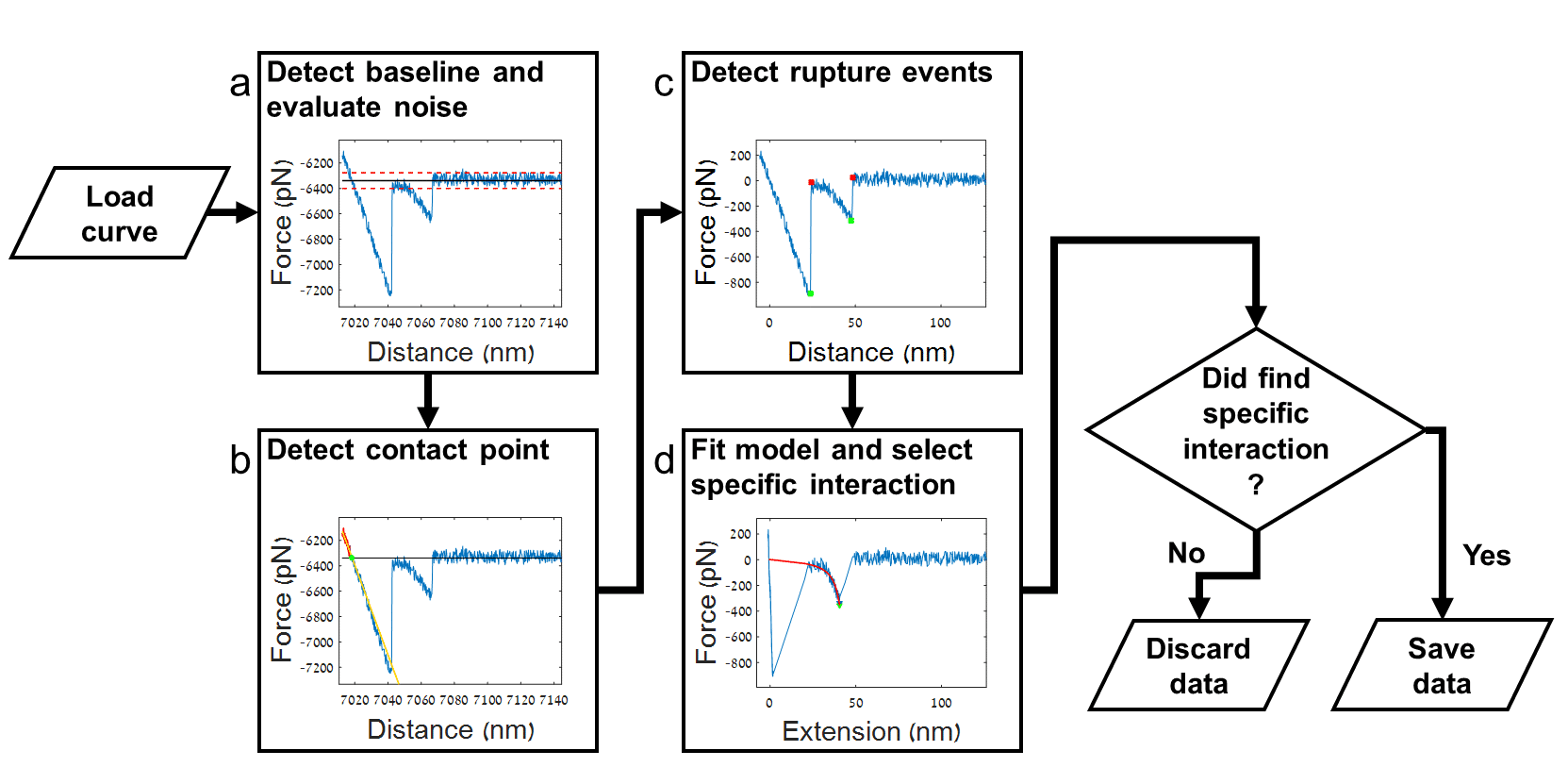
Duanis-Assaf, T., Razvag, Y., Reches, M., (2019) ForSDAT: An Automated Platform for Analyzing Force Spectroscopy Measurements, Anal. Methods, 11(37), 4709-4718
Duanis-Assaf, T., Razvag, Y., Reches M., ForSDAT: An automated platform for analyzing force spectroscopy measurements, provisional application
Powered by Eventact EMS