
Reduce food losses by developing DNA-based biosensor for a shelf-life prediction of the stored agriculture produce
2Agro-Nanotechnology and Advanced Materials Research Center, Volcani Institute, Israel
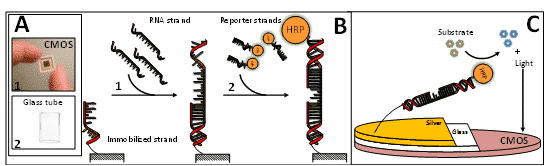
Postharvest fruit decay is caused by fungal pathogens and leads to major losses. In this study, specific mRNA sequences that are upregulated in the fungus Colletotrichum gloeosporioides during its quiescent stage in fruits were identified using a CMOS sensor. The identification process was based on a sandwich approach, where strands complementary to the C. gloeosporioides mRNA sequences (quiescent stage-specific) were immobilized on the CMOS surface and exposed to the target complementary reporter strands. In the presence of a target sequence, the reporter strand (linked to the enzyme horseradish peroxidase (HRP)) was left in the system and a measurable light signal was produced. The complementary strands are specifically annealed to the mRNA in the sample. The sensitivity of the technology was assessed by mRNA sequences isolated from C. gloeosporioides and identified as 10 nM RNA. The effect of the pathogenicity state on the sensor performance was also evaluated. The CMOS sensor could detect quiescent fungi, which are barely detectable by other means. Then, the proposed system was optimized by adding the metal-enhanced fluorescence (MEF) phenomenon to the measuring processes, while signal intensity was amplified by silver nanoislands deposited onto a glass surface. The photons from the light signal produced from the HRP enzyme and its substrate were coupled by silver nanoislands and enhanced. The implementation of the improved conditions increased the biosensor sensitivity to 3.3 nM. The obtained valuable information from this platform allows the early detection of RNA-markers of quiescent pathogenic fungi in agricultural produce, which will provide a smart and data-based decision-making tool to reduce postharvest losses.
Powered by Eventact EMS