THE STRUCTURAL BASIS FOR THE VIRULENCE AND HOST-PATHOGEN INTERACTIONS OF MICROBIAL AMYLOIDS
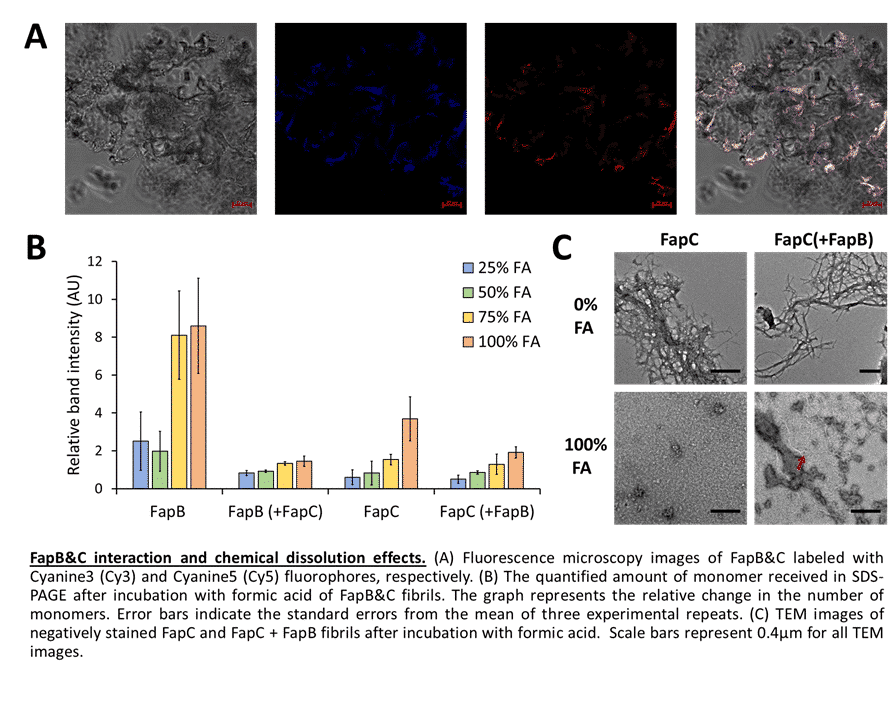
Amyloid study has advanced significantly over the past few decades, especially in eukaryotes where amyloids are thought to contribute to neurodegenerative and other systemic diseases; yet, amyloids are also found in many other organisms and can carry out various physiological processes. They are especially prevalent in microbes and were identified as key virulence factors, thus microbial amyloids represent attractive candidates for structural characterization aimed at discovering novel antimicrobial therapeutics. However, amyloids are challenging systems for biochemical and structural studies due to their polymeric arrangement and aggregative, polymorphic, and partially disordered nature. We have devoted much of our recent study efforts on three well-known families of amyloid proteins: Enterobacteria Curli, Pseudomonas Fap, and Candida Als. Our recent studies have resolved the structures, as well as evaluated the functional significance of amyloidogenic peptide segments within the adhesion protein Als5 of Candida albicans. Additionally, we investigated the interaction between FapB and FapC, two predominating amyloid proteins of Pseudomonas aeruginosa. We illustrated how they are affected by different growth conditions, like pH and temperature, and how their interactions tend to produce fibrils that are much more resistant to harsh environmental conditions. These results might suggest a different "distribution of labor" between the proteins than previously suggested. A detailed description of the structure and activity of microbial amyloid proteins will allow us to better deal with infections caused by these pathogens, and even repurpose their role to utilize to our benefit the potential inherent in them.