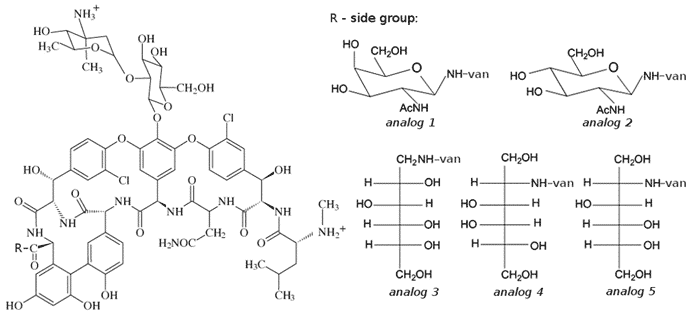
An extensive computational investigations have been performed to
examine the effects of the addition of 2-acetamido-2-deoxy-b-D-galactopyranosylamine (analog 1),
2-acetamido-2-deoxy-b-D-glucopyranosylamine
(analog 2), 1-amino-1-deoxy-D-glucitol (analog
3), 2-amino-2-deoxy-D-galactitol (analog 4)
and 2-amino-2-deoxy-D-glucitol (analog 5) to the C-terminal
amino acid group in the vancomycin.
All non standard groups have been parametrized for the gaff
and glycam06 force fields, and connected to the heptapeptide macrocyclic
vancomycin ring C-termini by a peptide bond. A pentapeptide cell wall precursor
mimic AcAla-D-iGlu-Lys-D-Ala-D-Ala has been added in the position known to form
active complex with vancomycin. To the computational system there has been also
added a periodical, pre-equilibrated water box, TIP3P model.
Every vancomycin
analog-peptidoglycan precursor complex has been optimized, submitted to the
isothermal-isobaric molecular dynamics in the AMBER package and then analyzed.
The analysis of overall RMSd changes, changes in position and interactions
involving modified part of vancomycin as well as comparative study of possible
interactions with cyclic and chain forms of modified groups are discussed.
Acknowledgments: This work was supported by the
European Union within the European Regional Development Fund (grant
UDA-POIG.01.01.02-14-102/09/03). The computational time in the Academic Computer Center in Gdansk CI
TASK, Poland is acknowledged.

