Protein-carbohydrate interactions typically exhibit high specificity and weak affinities toward their carbohydrate ligand. This low affinity is compensated in nature by the architecture of the protein, the host presenting the carbohydrate ligands in a multivalent manner on the cell or mucosal surface.1 In order to overcome the low binding affinities of monovalent glycosides, chemists have successfully conjugated synthetic oligosaccharides to a variety of multivalent scaffolds. Among the many platforms available, Quantum Dots (QDs) are particularly attractive materials that can be synthesized to emit light in well-separated ranges across the visible spectrum and their inherent electron density makes them ideal cellular markers for correlative light and electron microscopy.2
Glycan-QDs constitute a good bio-mimetic model of carbohydrate presentation at the cell surface and provide a powerful tool to screen for protein-carbohydrate interactions.3 To use these nanoparticles in biomedical applications, it is important to understand the parameters that control particle stability in physiological media and the effect that specific capping groups have on particle cellular uptake, localization and toxicity.
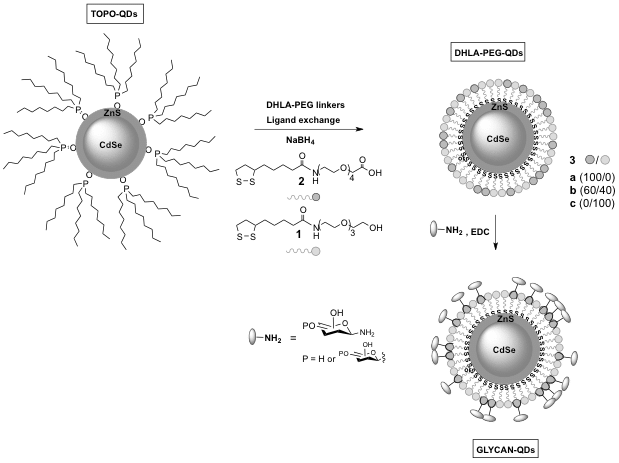
Herein we report a simple and convenient synthesis of sugar coated PEGylated CdSe/ZnS QDs with varying carbohydrate type and surface densities that can be used to study cellular uptake. Using HeLa and human corneal epithelial cells (Araki-Sasaki), we show that cellular uptake and intracellular localization is dependent on the type of sugar, while carbohydrate surface density has an impact on cellular uptake and toxicity. We show that certain glycan sequences can be used as a “Trojan Horse” to help internalize other moieties that would otherwise not be found within these cells.4
(1) Lundquist, J. J.; Toone, E. J. Chem Rev 2002, 102, 555.
(2) Nozik, A. J. et al. Chem Rev 2010, 110, 6873.
(3) Kikkeri, R. et al. J Am Chem Soc 2009, 131, 2110.
(4) Galan, M.C. et al. Submitted 2013

