Cancer is a heterogeneous and tissue specific disease. Consequently, it is suggested that tissue differentiation, mediated mostly by epigenetic modifications, could guide tissue specific susceptibility and protective mechanisms against cancer. In this work we established genome wide methylation profiles of various normal epithelial tissues and identified genes that are differentially methylated in breast epithelium. We further elaborated on TRIM29 (ATDC) regulation in various epithelial tissues and related cancers. The promoter of TRIM29 was hypo-methylated in normal breast and prostate epithelium and densely methylated in the other epithelial tissues tested. Accordingly, TRIM29 expression inversely correlated with promoter methylation. Review of the public genomic data bases TCGA and ENCODE confirmed these findings. Furthermore, methylation increased and expression decreased in breast and prostate carcinomas whereas the reverse was noted for multiple other carcinomas. Interestingly, TRIM29 regulation in breast tumors clustered according to the PAM50 subtypes. These findings suggest that TRIM29 has a multifunctional role in the differentiation of normal epithelial tissues and in the evolution of the respective cancers.
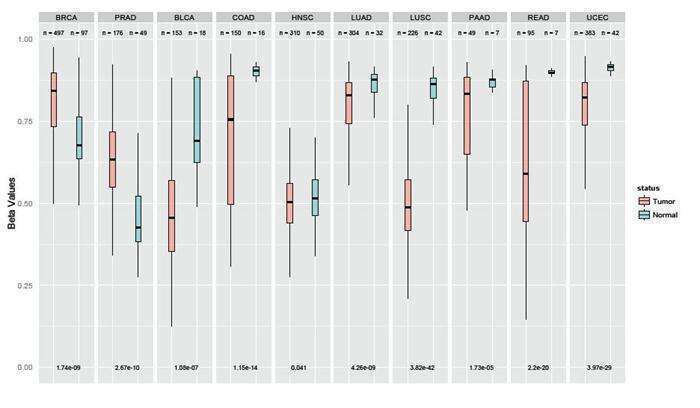
TRIM29 promoter methylation in normal and cancer tissues. Data obtained from the TCGA data base. BRCA=Breast, PRAD=Prostate, BLCA=bladder COAD=Colon, HNSC=Head&Neck, LUAD=Lung adeno, LUSC=Lung squamous, PAAD=Pancreas, READ=Renal, UCEC=Uterine-Cervix.