Background: In 1996, the first summary of molecular genetics of familial hypercholesterolemia (FH) in Israel was published (Reshef A, Hum Gen, 1996). Since, new technology for genotyping became available making genetic diagnosis easier more accurate, and more comprehensive. We performed systematic genotyping index cases of 68 pedigrees to reanalyze the profile of mutation among Israeli FH.
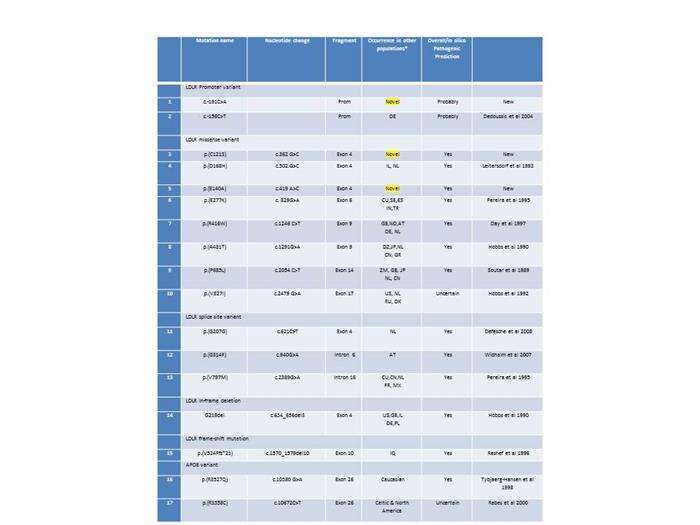
Methods and Results: LRLR and APOB genes were sequenced in 68 individuals using sanger sequencing. Mutations of the LDLR and APOB gene were found in twenty-three patients of the Israeli cohort which comprise 34% of the Israeli total. Among the twenty-three patients, there were seventeen different of LDLR mutations and the p.(R3527Q) and p.(R3558C) APOB mutations (table 1). Three variants, two in exon 4 and one in the promoter region were not reported before. one patient carried the common APOB p.(R3527Q) mutation. 70% of out cohort were negative for LDL or APOB mutation. It was hypothesized that FH can be caused by an accumulation of LDL-C raising alleles having small contributive effect (Talmud et al., 2013). Talmud and colleagues developed a gene score method derived from twelve common LDL-C raising SNPs in eleven genes. The mean poly gene score was higher among mutation negative FH patients compared with a European control population.
Conclusion: By using direct sequencing we identified muations in 30% of an ungenotyhped Israeli FH cohort. In these we found 3 novel LDLR mutation and new APOB mutation in the Israeli population. In the remaining mutation free cohort, the Talmud LDLc polygene score was higher that in controls. We conclude that mutation causing FH can be identified in 30% of FH patients. These data can help design future strategy for early screening for FH in our population.