
AUTOMATED ANALYSIS OF FLUORESCENCE KINETICS IN SINGLE MOLECULE LOCALIZATION MICROSCOPY DATA REVEALS PROTEIN STOICHIOMETRY
2Institute of Physical and Theoretical Chemistry, Goethe University Frankfurt, Frankfurt, Germany
3Electrical Engineering, Technion - Israel Institute of Technology, Haifa, Israel
The molecular organization of protein complexes holds information about their function and about changes in the cell state. Specifically, some proteins might change their oligomerization level to initiate processes inside cells, such as cellular motility, differentiation, etc. Super-resolution microscopy has enabled researchers to observe single proteins with high precision beyond the diffraction limit. However, the improvement in the resolution is not sufficient to differentiate between proteins within the same cluster, therefore, quantification of protein oligomerization level remains a challenge. In this work, we present a Quantitative Algorithm for Fluorescent Kinetics Analysis (QAFKA), that serves as a fully automated workflow to quantify oligomerization level in protein populations. QAFKA processes single molecule localization microscopy (SMLM) data by extracting fluorophore “blinking” events and analyzing their temporal characteristics. The algorithm is composed of three main blocks (see figure): super-resolution localization of emission events, blinking kinetics feature extraction and a deep neural network based estimator that reports the ratios of different cluster populations. We demonstrate molecular quantification of protein monomers and dimers in simulated and experimental SMLM data. We show QAFKA robustness to various parameters such as photobleaching, labeling density, etc. Furthermore, QAFKA is capable to accurately report monomer/ dimer equilibrium when presented with relatively short experiments compared to other state-of-the-art methods. Finally, we demonstrate that QAFKA capability to accurately report quantitative information on the monomer/ dimer equilibrium of membrane receptors in single immobilized cells, opening the door to single-cell single-protein analysis.
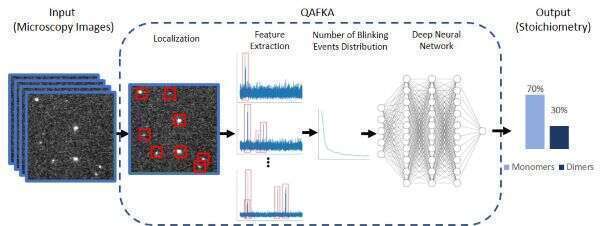
Figure: Algorithm block diagram of QAFKA. The input is a photoactivated localization microscopy (PALM) movie containing stochastic blinking events of single molecules. The automated analysis algorithm localizes each cluster, extracts relevant emission features, and finally predicts the stoichiometry of monomers and dimers using a deep neural network based estimator.