
HOW cryo-EM ILLUMINATES THE UNIQUE TOPOLOGY AND PRE-LOADED RECEPTOR PRESENT WITHIN THE LASSA VIRUS’ SPIKE COMPLEX
One of the most clinically relevant steps in the life cycle of pathogenic viruses is their cell entry. Therefore, it is critical to obtain detailed molecular-level descriptions for such infection mechanisms that pathogenic viruses use. The Lassa virus, a lethal member of the Arenaviridae family, infects thousands of people each year throughout West Africa. This virus utilizes its spike protein to engage with its endogenous host receptor, matriglycan, a post-translational modification on α-dystroglycan (α-DG). Despite extensive past research, many open questions remain regarding the mechanism that allows the Lassa virus to enter cells. It is unknown, for example, how matriglycan is recognized. We do not know how the spike complex can withstand harsh environmental conditions, as previous observations suggested that portions of this spike are very labile. Moreover, arenaviruses have a stable signal peptide (SSP), which makes up an integral part of the spike and is critical for its function, yet the structure and topology of the SSP in the membrane are not known.
We therefore set out to address these open questions by solving the structure of the native, full-length trimeric spike, solubilized directly from membranes. Our method of choice was single-particle cryo-electron microscopy (cryo-EM). Solving the structure of the spike was a challenging endeavor, with numerous obstacles in each and every step of the way. Using a trial-and-error approach, we managed to develop a production and purification scheme that allowed us to solubilize the spike in detergent while keeping its native conformation. Preparing grids with sufficient particle density and collecting single-particle data with adequate angular coverage also turned out to be a difficult task that was ultimately addressed using a graphene-oxide coating and additional tweaks in the purification procedure. Eventually, we were able to reconstruct a C3-symmetric density map for the Lassa spike complex at a Gold-Standard Fourier Shell Correlation (FSC) resolution of 3.3 Å, which allowed us to solve the structure. The structure of the spike revealed that the arenaviral SSP exists in an unexpected topology in the membrane, which further indicates a conformational switch by one of its transmembrane helices. At both the membrane-proximal region and the apex of the spike, we discovered domain swapping interactions that rigidify the spike, rationalizing the previously unexplained stability of the complete spike complex.
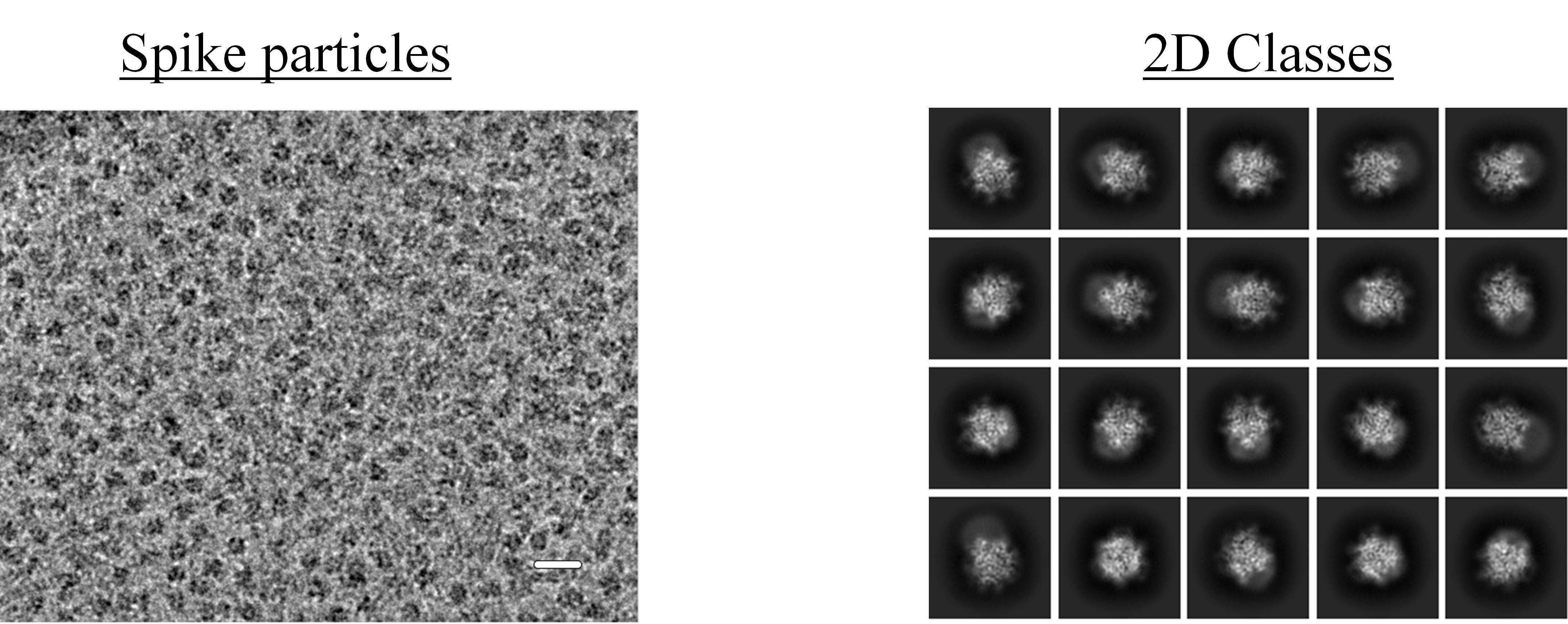
Figure 1: (Left) Representative cryo-EM micrograph of Lassa spike particles, obtained from the Talos Arctica. The scale bar is 200 Å. (Right) 2D classes obtained after selecting and averaging particles from data collection on the Titan Krios G3i.
We further reconstructed a C1-symmetric density map, which allowed us to discover a non-symmetric density blob to which we were able to fit a linear chain of matriglycan. Modeling of matriglycan, which was corroborated using biochemical analyses, allowed us to elucidate for the first time how the Lassa virus attaches to its host cell. This is a groundbreaking achievement that answered a decades-long open question in the field and further provided a molecular-level rationalization for a large body of biochemical data gathered over the years. Moreover, discovering a non-covalently linked fragment of matriglycan bound by the spike complex of the Lassa virus is by itself a startling finding. Not only does it reveal a previously unknown ability of cells to produce such a substance, but it also suggests that newly formed spike complexes of the Lassa virus will not be able to engage with their cellular receptor since they are blocked. This is a seminal finding as it provides a mechanism for another step in the life cycle of the Lassa virus, which is the egress from its host cell. This mechanism for egress is likely also shared by other α-DG-tropic arenaviruses.
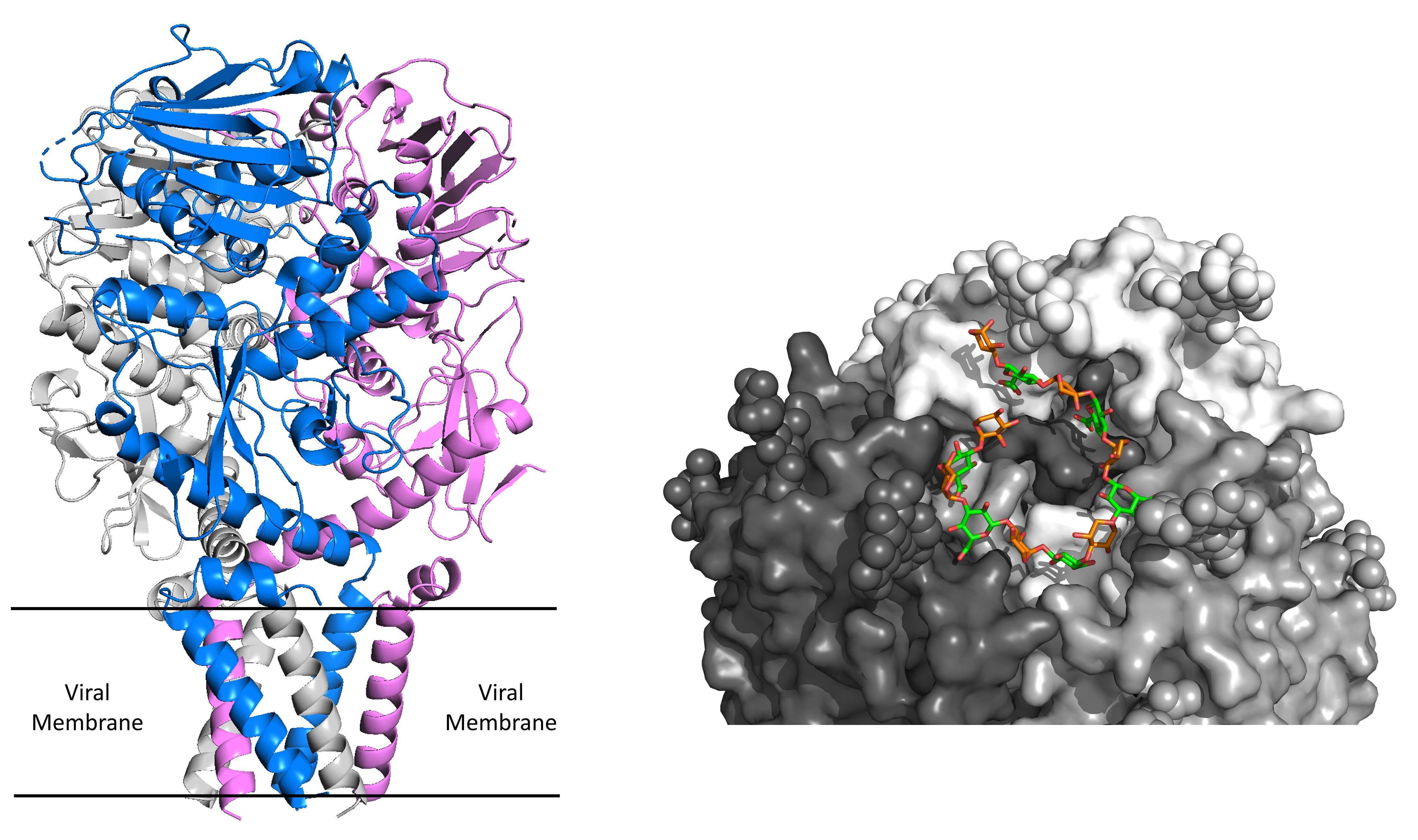
Figure 2: (Left) Structure of the native, full-length Lassa spike complex. Each protomer of the trimer is shown in a different color. The location of the six transmembrane helices relative to the viral membrane is indicated. (Right) A membrane-distal view of the soluble matriglycan fragment (green & orange) bound to the active site of the Lassa spike.
Collectively, we used an electron microscopy approach to provide novel structural information that answered many important open questions in our field and further led to the discovery of new and exciting mechanisms that the Lassa virus is utilizing to infect cells. These data, that were recently published in a Nature paper, may facilitate the development of new therapeutic approaches for fighting the notorious Lassa virus.